AbEpiTope-1.0: Improved antibody target prediction by use of AlphaFold and inverse folding
- PMID: 40512857
- PMCID: PMC12165000
- DOI: 10.1126/sciadv.adu1823
AbEpiTope-1.0: Improved antibody target prediction by use of AlphaFold and inverse folding
Abstract
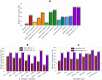
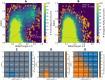
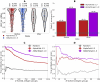
B cell epitope prediction tools are crucial for designing vaccines and disease diagnostics. However, predicting which antigens a specific antibody binds to and their exact binding sites (epitopes) remains challenging. Here, we present AbEpiTope-1.0, a tool for antibody-specific B cell epitope prediction, using AlphaFold for structural modeling and inverse folding for machine learning models. On a dataset of 1730 antibody-antigen complexes, AbEpiTope-1.0 outperforms AlphaFold in predicting modeled antibody-antigen interface accuracy. By creating swapped antibody-antigen complex structures for each antibody-antigen complex using incorrect antibodies, we show that predicted accuracies are sensitive to antibody input. Furthermore, a model variant optimized for antibody target prediction-differentiating true from swapped complexes-achieved an accuracy of 61.21% in correctly identifying antibody-antigen pairs. The tool evaluates hundreds of structures in minutes, providing researchers with a resource for screening antibodies targeting specific antigens. AbEpiTope-1.0 is freely available as a web server and software.
Figures


Similar articles
-
Mining for the antibody-antigen interacting associations that predict the B cell epitopes.BMC Struct Biol. 2010 May 17;10 Suppl 1(Suppl 1):S6. doi: 10.1186/1472-6807-10-S1-S6. BMC Struct Biol. 2010. PMID: 20487513 Free PMC article.
-
Antibody-protein interactions: benchmark datasets and prediction tools evaluation.BMC Struct Biol. 2007 Oct 2;7:64. doi: 10.1186/1472-6807-7-64. BMC Struct Biol. 2007. PMID: 17910770 Free PMC article.
-
Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes.Front Immunol. 2019 Feb 26;10:298. doi: 10.3389/fimmu.2019.00298. eCollection 2019. Front Immunol. 2019. PMID: 30863406 Free PMC article.
-
Discovery of protective B-cell epitopes for development of antimicrobial vaccines and antibody therapeutics.Immunology. 2014 May;142(1):1-23. doi: 10.1111/imm.12213. Immunology. 2014. PMID: 24219801 Free PMC article. Review.
-
An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction.J Immunol Res. 2016;2016:6760830. doi: 10.1155/2016/6760830. Epub 2016 Dec 29. J Immunol Res. 2016. PMID: 28127568 Free PMC article. Review.
References
-
- Kozlova E. E. G., Cerf L., Schneider F. S., Viart B. T., Nguyen C., Steiner B. T., de Almeida Lima S., Molina F., Duarte C. G., Felicori L., Chávez-Olórtegui C., Machado-de-Ávila R. A., Computational B-cell epitope identification and production of neutralizing murine antibodies against Atroxlysin-I. Sci. Rep. 8, 14904 (2018). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

