Amplicon sequencing of pasteurized retail dairy enables genomic surveillance of H5N1 avian influenza virus in United States cattle
- PMID: 40513042
- PMCID: PMC12165699
- DOI: 10.1371/journal.pone.0325203
Amplicon sequencing of pasteurized retail dairy enables genomic surveillance of H5N1 avian influenza virus in United States cattle
Abstract
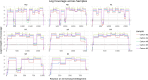
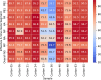
Highly pathogenic avian influenza (HPAI) viruses with H5 hemagglutinin (HA) genes (clade 2.3.4.4b) are causing an ongoing panzootic in wild birds. Circulation of these viruses is associated with spillover infections in multiple species of mammals, including a large, unprecedented outbreak in American dairy cattle. Before widespread on-farm testing, there was an unmet need for genomic surveillance. Infected cattle can shed high amounts of HPAI H5N1 viruses in milk, allowing detection in pasteurized retail dairy samples. Over a 2-month sampling period in one Midwestern city, we obtained dairy products processed in 20 different states. Here we demonstrate that a tiled-amplicon sequencing approach produced over 90% genome coverage at greater than 20x depth from 5 of 13 viral RNA positive samples, with higher viral copies corresponding to better sequencing success. The sequences clustered phylogenetically within the rest of the cattle outbreak sequences reported. A combination of RT-qPCR testing and sequencing from retail dairy products can be a useful component of a One Health framework for responding to the avian influenza outbreak in cattle.
Copyright: © 2025 Lail et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: AJL has received travel funding from Oxford Nanopore Techologies to present this work at a conference. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

