DHODH Blockade Induces Ferroptosis in Neuroblastoma by Modulating the Mevalonate Pathway
- PMID: 40513779
- PMCID: PMC12275935
- DOI: 10.1016/j.mcpro.2025.101014
DHODH Blockade Induces Ferroptosis in Neuroblastoma by Modulating the Mevalonate Pathway
Abstract
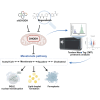
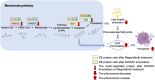
Neuroblastoma is the most common heterogeneous solid tumor in children, and current treatment options remain limited, especially for high-risk patients. Previous studies have identified dihydroorotate dehydrogenase (DHODH), a key enzyme in pyrimidine synthesis, as a potential therapeutic target in cancer. However, none of the existing FDA-approved DHODH inhibitors have shown effective inhibition of neuroblastoma cell growth. To address this challenge, we employed virtual screening to discover potential DHODH-targeting drugs, identifying Regorafenib as a promising candidate. Regorafenib significantly inhibited neuroblastoma growth in both neuroblastoma cells and patient-derived organoids. To unravel the underlying molecular mechanisms, we conducted Tandem Mass Tag (TMT)-based quantitative proteomics using LC-MS/MS. Our proteomic profiling revealed substantial regulation of lipid metabolism proteins, specifically those in the mevalonate pathway, correlating with ferroptosis induction. Further analysis showed that DHODH inhibition led to a reduction in total cholesterol, cholesterol esters, disrupted lipid droplet formation, and significantly decreased the expression of Squalene Epoxidase (SQLE), a key enzyme in lipid metabolism. Notably, we also observed an increase in nuclear SQLE expression following DHODH inhibition. In summary, our study highlights DHODH blockade as a novel approach to induce ferroptosis through lipid metabolism reprogramming, underscoring DHODH as a viable therapeutic target for neuroblastoma treatment. These insights open new avenues for metabolism-based interventions in aggressive pediatric cancers.
Keywords: DHODH; ferroptosis; lipid metabolism; mevalonate pathway; neuroblastoma; proteomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
-
- Brodeur G.M. Neuroblastoma: biological insights into a clinical enigma. Nat. Rev. Cancer. 2003;3:203–216. - PubMed
-
- Gatta G., Botta L., Rossi S., Aareleid T., Bielska-Lasota M., Clavel J., et al. Childhood cancer survival in Europe 1999-2007: results of EUROCARE-5--a population-based study. Lancet Oncol. 2014;15:35–47. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

