Extracellular DNA filaments associated with surface polysaccharide II give Clostridioides difficile biofilm matrix a network-like structure
- PMID: 40514355
- PMCID: PMC12166085
- DOI: 10.1038/s41522-025-00751-5
Extracellular DNA filaments associated with surface polysaccharide II give Clostridioides difficile biofilm matrix a network-like structure
Abstract
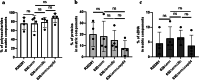
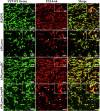
Clostridioides difficile is an anaerobic, spore-forming, Gram-positive bacterium, and a leading cause of healthcare-associated intestinal infections. Recurrences occur frequently, most of them being relapses. Apart from spores, C. difficile biofilm is hypothesized as a reservoir for relapses. Thus, increased knowledge on in vitro biofilm formation and characteristics is required. We finely characterized the matrix components in 4 C. difficile strains. Confocal microscopy revealed for the first time the presence of eDNA filaments connecting bacteria, with a spider's web-like organization. Biofilm disruption with DNase I suggests that eDNA, even in low abundance, plays a key role in the biofilm scaffold, maintaining biofilm cohesion by connecting bacteria. Observation of strong overlapping staining, particularly in the highest biofilm-producing strain tested between eDNA and polysaccharide II or lipoprotein CD1687, suggests that interactions between these components may enhance biofilm cohesion. Whereas autolysis does not appear to be a major way of matrix component release under our conditions, eDNA was sometimes associated with lipidic round shapes that can evoke vesicle structures. Together, these results suggest that the bacterial aggregation and structuring of the C. difficile biofilm involve several components of the matrix, including eDNA, interacting with each other to build the scaffold of biofilm.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
The cell wall lipoprotein CD1687 acts as a DNA binding protein during deoxycholate-induced biofilm formation in Clostridioides difficile.NPJ Biofilms Microbiomes. 2023 May 11;9(1):24. doi: 10.1038/s41522-023-00393-5. NPJ Biofilms Microbiomes. 2023. PMID: 37169797 Free PMC article.
-
Spore formation and toxin production in Clostridium difficile biofilms.PLoS One. 2014 Jan 30;9(1):e87757. doi: 10.1371/journal.pone.0087757. eCollection 2014. PLoS One. 2014. PMID: 24498186 Free PMC article.
-
Extracellular DNA, cell surface proteins and c-di-GMP promote biofilm formation in Clostridioides difficile.Sci Rep. 2021 Feb 5;11(1):3244. doi: 10.1038/s41598-020-78437-5. Sci Rep. 2021. PMID: 33547340 Free PMC article.
-
Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence.PLoS Pathog. 2021 Sep 9;17(9):e1009817. doi: 10.1371/journal.ppat.1009817. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499698 Free PMC article. Review.
-
Type IV pili are involved in phenotypes associated with Clostridioides difficile pathogenesis.Crit Rev Microbiol. 2024 Nov;50(6):1011-1019. doi: 10.1080/1040841X.2023.2235002. Epub 2023 Jul 15. Crit Rev Microbiol. 2024. PMID: 37452617 Review.
References
-
- Le Monnier, A. et al. Hospital cost of Clostridium difficile infection including the contribution of recurrences in French acute-care hospitals. J. Hosp. Infect.91, 117–122 (2015). - PubMed
-
- Reigadas, E., Vázquez-Cuesta, S. & Bouza, E. Economic burden of Clostridioides difficile Infection in European Countries. Adv. Exp. Med. Biol.1435, 1–12 (2024). - PubMed
-
- Heise, J., Witt, P., Maneck, C., Wichmann-Schauer, H. & Maurischat, S. Prevalence and phylogenetic relationship of Clostridioides difficile strains in fresh poultry meat samples processed in different cutting plants. Int J. Food Microbiol.339, 109032 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

