Cathepsin L-dependent positive selection shapes clonal composition and functional fitness of CD4+ T cells
- PMID: 40514418
- PMCID: PMC12208919
- DOI: 10.1038/s41590-025-02182-y
Cathepsin L-dependent positive selection shapes clonal composition and functional fitness of CD4+ T cells
Abstract
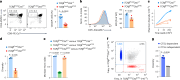
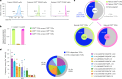
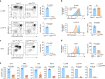
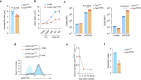
The physiological significance of thymic positive selection and its reliance on a single stromal cell type, cortical thymic epithelial cells, remain incompletely understood. The lysosomal cysteine protease cathepsin L (CTSL) has been implicated in generating major histocompatibility complex class II-bound peptides in cortical thymic epithelial cells for efficient CD4+ T cell differentiation. Here, we addressed the extent and nature of the CD4+ T cell repertoire changes associated with CTSL deficiency. In the absence of CTSL, a highly selective loss of T cell receptors resulted in a markedly reduced repertoire diversity. However, a similarly large proportion of nominally 'CTSL-independent' T cell receptors were retained. Clones representative of the second category experienced weaker positive selection signals in the absence of CTSL, which were sufficient for further maturation yet imprinted aberrant responsiveness to agonist stimulation and impaired homeostatic behavior. Together, these findings demonstrate that CTSL is crucial for both shaping full repertoire diversity and optimizing CD4+ T cell functionality.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









Similar articles
-
Cortical Thymic Epithelial Cells: Key Developers of the Code of T-Cell Selection and TCR Repertoire Diversity.Immunol Rev. 2025 Jul;332(1):e70049. doi: 10.1111/imr.70049. Immunol Rev. 2025. PMID: 40558027 Review.
-
Integrin CD103 reveals a distinct developmental pathway of autoreactive thymocytes in TCR transgenic mice.Nat Commun. 2025 Jul 18;16(1):6627. doi: 10.1038/s41467-025-61922-8. Nat Commun. 2025. PMID: 40681578 Free PMC article.
-
LAMP2 regulates autophagy in the thymic epithelium and thymic stroma-dependent CD4 T cell development.Autophagy. 2023 Feb;19(2):426-439. doi: 10.1080/15548627.2022.2074105. Epub 2022 May 19. Autophagy. 2023. PMID: 35535798 Free PMC article.
-
CD6 regulates CD4 T follicular helper cell differentiation and humoral immunity during murine coronavirus infection.J Virol. 2025 Jan 31;99(1):e0186424. doi: 10.1128/jvi.01864-24. Epub 2024 Dec 16. J Virol. 2025. PMID: 39679790 Free PMC article.
-
Structured treatment interruptions (STI) in chronic unsuppressed HIV infection in adults.Cochrane Database Syst Rev. 2006 Jul 19;2006(3):CD006148. doi: 10.1002/14651858.CD006148. Cochrane Database Syst Rev. 2006. PMID: 16856117 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

