Cell state-specific cytoplasmic density controls spindle architecture and scaling
- PMID: 40514430
- PMCID: PMC12173940
- DOI: 10.1038/s41556-025-01678-x
Cell state-specific cytoplasmic density controls spindle architecture and scaling
Abstract
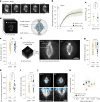
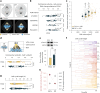
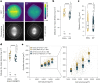
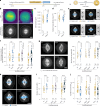
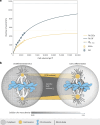
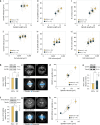
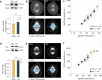
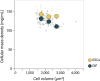
Mitotic spindles are dynamically intertwined with the cytoplasm they assemble in. How the physicochemical properties of the cytoplasm affect spindle architecture and size remains largely unknown. Using quantitative biochemistry in combination with adaptive feedback microscopy, we investigated mitotic cell and spindle morphology during neural differentiation of embryonic stem cells. While tubulin biochemistry and microtubule dynamics remained unchanged, spindles changed their scaling behaviour; in differentiating cells, spindles were considerably smaller than those in equally sized undifferentiated stem cells. Integrating quantitative phase imaging, biophysical perturbations and theory, we found that as cells differentiated, their cytoplasm became more dilute. The concomitant decrease in free tubulin activated CPAP (centrosomal P4.1-associated protein) to enhance the centrosomal nucleation capacity. As a consequence, in differentiating cells, microtubule mass shifted towards spindle poles at the expense of the spindle bulk, explaining the differentiation-associated switch in spindle architecture. This study shows that cell state-specific cytoplasmic density tunes mitotic spindle architecture. Thus, we reveal physical properties of the cytoplasm as a major determinant in organelle size control.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Similar articles
-
SART1 uniquely localizes to spindle poles forming a SART1 cap and promotes spindle pole assembly.J Biol Chem. 2025 Jun;301(6):108561. doi: 10.1016/j.jbc.2025.108561. Epub 2025 May 2. J Biol Chem. 2025. PMID: 40320072 Free PMC article.
-
The cell cycle oscillator and spindle length set the speed of chromosome separation in Drosophila embryos.Curr Biol. 2025 Feb 3;35(3):655-664.e3. doi: 10.1016/j.cub.2024.11.046. Epub 2025 Jan 9. Curr Biol. 2025. PMID: 39793565
-
Plasticity of the mitotic spindle in response to karyotype variation.Curr Biol. 2024 Aug 5;34(15):3416-3428.e4. doi: 10.1016/j.cub.2024.06.058. Epub 2024 Jul 22. Curr Biol. 2024. PMID: 39043187 Free PMC article.
-
Regulation of microtubule growth rates and their impact on chromosomal instability.Cell Cycle. 2024 Jul-Aug;23(13-16):872-891. doi: 10.1080/15384101.2025.2485842. Epub 2025 Apr 22. Cell Cycle. 2024. PMID: 40260826 Review.
-
Mechanisms of Mitotic Spindle Assembly.Annu Rev Biochem. 2016 Jun 2;85:659-83. doi: 10.1146/annurev-biochem-060815-014528. Epub 2016 Apr 21. Annu Rev Biochem. 2016. PMID: 27145846 Free PMC article. Review.
Cited by
-
Cytoplasmic control of spindle architecture.Nat Cell Biol. 2025 Jun;27(6):884-885. doi: 10.1038/s41556-025-01686-x. Nat Cell Biol. 2025. PMID: 40527946 No abstract available.
References
-
- Thompson, D. W. On Growth and Form (Cambridge Univ. Press, 1992).
-
- Greenan, G. et al. Centrosome size sets mitotic spindle length in Caenorhabditis elegans embryos. Curr. Biol.20, 353–358 (2010). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

