Synergistic effect of electrolyzed oxidized water (EO) and peroxyacetic acid on plasmid-mediated quinolone resistance genes of Pseudomonas aeruginosa
- PMID: 40514545
- PMCID: PMC12165879
- DOI: 10.1007/s11274-025-04384-w
Synergistic effect of electrolyzed oxidized water (EO) and peroxyacetic acid on plasmid-mediated quinolone resistance genes of Pseudomonas aeruginosa
Abstract
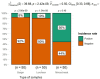
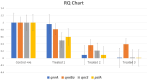
Pseudomonas aeruginosa displays major biofilm formation, food spoilage, and significant losses for the food industry. It extremely exhibits inherent resistance to various antibiotics, including quinolones, which can spread via plasmids. The present study investigates the prevalence of biofilm-forming P. aeruginosa in meat products and assesses its antibiotic resistance patterns. Evaluating the effectiveness of electrolyzed-oxidized water (EOW) and per-acetic acid (PAA) alone or in a combination against P. aeruginosa biofilm formed on stainless steel (SS) surfaces. Besides, the effect on quinolone-resistance (qnr) gene expression level using real-time PCR. P. aeruginosa was isolated from 24.6% of the total tested samples (37/150), with a significant variation observed regarding the contamination levels of the tested products. 36.3% of the isolates demonstrated robust biofilm production, with 82.3% displaying multi-drug resistance (MDR). Isolates revealed complete susceptibility to amikacin (100%) and gentamycin (82%). Quinolone resistance was observed in 23% and 17% of the isolates for ciprofloxacin and levofloxacin, respectively. 80% (4/5) among the confirmed isolates were enclosed the plasmid qnr genes. The genes pslA and gyrA were detected in 40% and 60%, respectively. EOW, particularly when combined with PAA, reveals strong antibacterial activity against P. aeruginosa and greatly decreases its count. Moreover, it can eliminate its biofilm within 20 min of exposure and significantly decrease the expression levels of quinolone-resistant genes. In conclusion, PAA and EOW are effective agents for biofilm control on SS surfaces, particularly when used in combined form, and could be more effective in combating resistant bacterial infections and controlling the spread of PMQR.
Keywords: P. aeruginosa; Per-acetic acid; Plasmid-mediated quinolone resistance (PMQR) genes. Electrolyte oxidized water (EOW).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Abayneh M, Tesfaw G, Woldemichael K, Yohannis M, Abdissa A (2019) Assessment of extended-spectrum β-lactamase (ESBLs) - producing Escherichia coli from minced meat of cattle and swab samples and hygienic status of meat retailer shops in Jimma town, Southwest Ethiopia. BMC Infect Dis No 19(1):897. 10.1186/s12879-019-4554-6 - DOI - PMC - PubMed
-
- Abdallah M, Khelissa O, Ibrahim A, Benoliel C, Heliot L, Dhulster P, Chihib N-E (2015) Impact of growth temperature and surface type on the resistance of Pseudomonas aeruginosa and Staphylococcus aureus biofilms to disinfectants. Int J Food Microbiol No 214:38–47. 10.1016/j.ijfoodmicro.2015.07.022 - DOI - PubMed
-
- Abdelraheem WM, Abdelkader AE, Mohamed ES, Mohammed MS (2020) Detection of biofilm formation and assessment of biofilm genes expression in different Pseudomonas aeruginosa clinical isolates. Meta Gene No 23:100646. 10.1016/j.mgene.2020.100646 - DOI
-
- Abed W, Kareem S (2021) Molecular detection of GyrA and MexA genes in Pseudomonas aeruginosa. Mol Biology Rep No 48. 10.1007/s11033-021-06820-0 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

