Comprehensive analysis of molecular markers linked to antimalarial drug resistance in Plasmodium falciparum in Northern, Northeastern and Eastern Uganda
- PMID: 40514714
- PMCID: PMC12164153
- DOI: 10.1186/s12936-025-05439-x
Comprehensive analysis of molecular markers linked to antimalarial drug resistance in Plasmodium falciparum in Northern, Northeastern and Eastern Uganda
Abstract
Background: In Uganda, antimalarial resistance in Plasmodium falciparum poses serious public health and treatment challenges. Globally, recent data have highlighted the roles of following genes in malaria resistance: Plasmodium falciparum dihydrofolate reductase (Pfdhfr), Plasmodium falciparum dihydropteroate synthetase (Pfdhps), Plasmodium falciparum chloroquine resistance transporter (Pfcrt), Plasmodium falciparum multidrug resistance gene 1 (Pfmdr1), and Plasmodium falciparum K13 propeller domain (Pfk13). This study investigated the prevalence and characteristics of P. falciparum molecular markers linked to antimalarial resistance in Northern, Northeastern, and Eastern Uganda.
Methods: This cross-sectional study collected 200 dried blood samples from children (2 months to 12 years) in Northern, Eastern, and Northeastern Uganda. Samples were from malaria-positive cases confirmed by rapid diagnostic tests and microscopy. Genomic DNA was extracted from these samples and analysed using Molecular Inversion Probes to detect Plasmodium falciparum genetic mutations. The sequencing was performed on the Illumina MiSeq platform, and raw data was organized and analysed with MIPTools software.
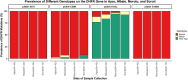
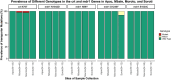
Results: The study sequenced over 50% of the samples at each site as follows: Apac 87.7% (43/49), Moroto 68.0% (34/50), Soroti 65.0% (13/20) and Mbale 53.1% (43/81). The Pfk13 A675V and C469Y mutations varied from 0 to 23.3% and 8.3-14.3%, in four sites, with consistently low prevalence in Apac. The Pfdhfr N51I and S108N mutations were fixed in all districts, while C59R was fixed in Moroto and nearing fixation (92-97%) in other regions. The emerging I164L mutation ranged from 1 to 10% in all sites. The Pfdhps A437G and K540E mutations were fixed in Soroti, with 3-5% wild-type prevalence in other sites. The A581G mutation showed 2.3% mixed genotypes in Mbale only. The Pfcrt K76T was predominantly wild type, except for 5% mutants in Mbale and Moroto. The pfmdr1 N86Y were wild type across all districts, except for 15% mixed genotypes in Soroti.
Conclusion: This study reveal rising partial artemisinin resistance and widespread antifolate resistance surpassing WHO thresholds in Northern, Northeastern, and Eastern Uganda. Emerging super-resistant parasites pose a serious threat to malaria control, necessitating urgent enhanced surveillance and alternative treatment strategies.
Keywords: Pfcrt; Pfdhfr; Pfdhps Pfmdr1; Pfk13; Antimalarial resistance; Uganda.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: A detailed protocol was developed beforehand and approved by the Mbale Regional Referral Hospital Research Ethics Committee [(MRRH-REC), MRRHREC-OUT-011/2020] and Uganda National Council for Science and Technology [(UNCST), HS3725ES]. Ethical considerations were adhered to during the study process. The study conformed to the provisions of ethical standards in Uganda. Consent for publication: The Mbale Clinical Research Institute (MCRI, www.mcri.ac.ug ), a research entity affiliated with the Uganda National Health Research Organization, approved the publication of this manuscript. Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials