Mitochondrial sirtuin 4 shapes the intestinal microbiota of Drosophila by controlling lysozyme expression
- PMID: 40514757
- PMCID: PMC12166577
- DOI: 10.1186/s42523-025-00431-x
Mitochondrial sirtuin 4 shapes the intestinal microbiota of Drosophila by controlling lysozyme expression
Abstract
Background: Sirtuins are deacetylases that are highly conserved throughout the animal kingdom. They act as metabolic sensors that coordinate cellular responses, allowing an adapted response to various stressors. Epithelial cells, especially those of the intestine, are directly exposed to a wide range of stressors. Together with the microbiota, they form a complex ecosystem with mutual influences. The significance of sirtuins in this complex system is still waiting to be clarified.
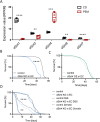
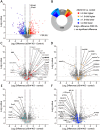
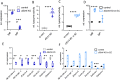
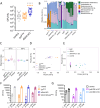
Results: Here, we show that a protein-restricted diet strongly increases the intestinal expression of sirtuin 4 (dSirt4), the only mitochondrial sirtuin in Drosophila. To elucidate the effects of deregulated dSirt4 expression in the intestine, we analyzed dSirt4 knockout flies. These flies showed substantial changes in their intestinal proteome and physiological properties. One of the most striking effects was the strong induction of lysozymes in the intestine, with a corresponding increase in lysozyme activity. This effect was organ-autonomous, as it was also observed in flies with dSirt4 knocked out only in intestinal enterocytes. The significant increase in lysozyme abundance in response to tissue-specific dSirt4 knockdown did not reduce the total number of bacteria in the intestine. However, it did affect the microbiota composition by reducing the number of gram-positive bacteria. This effect on microbiota composition can be attributed to dSirt4-dependent lysozyme expression, which is absent in a lysozyme-deficient background. dSirt4 knockout in the enterocytes shortened the lifespan of the flies, as did ectopic lysozyme overexpression in the enterocytes.
Conclusions: The only mitochondrial sirtuin in Drosophila, dSirt4, is induced by dietary stress in intestinal epithelial cells, which directly regulates the lysozyme activity of these cells. We could associate this altered lysozyme activity with a shift in the microbiota composition, demonstrating a direct link between stress, nutrition, and the host's microbiota regulation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Loss of mitochondrial SIRT4 shortens lifespan and leads to a decline in physical activity.J Biosci. 2018 Jun;43(2):243-247. J Biosci. 2018. PMID: 29872013
-
Intestinal lysozyme1 deficiency alters microbiota composition and impacts host metabolism through the emergence of NAD+-secreting ASTB Qing110 bacteria.mSystems. 2024 Mar 19;9(3):e0121423. doi: 10.1128/msystems.01214-23. Epub 2024 Feb 16. mSystems. 2024. PMID: 38364095 Free PMC article.
-
Drosophila Antimicrobial Peptides and Lysozymes Regulate Gut Microbiota Composition and Abundance.mBio. 2021 Aug 31;12(4):e0082421. doi: 10.1128/mBio.00824-21. Epub 2021 Jul 13. mBio. 2021. PMID: 34253067 Free PMC article.
-
Hydrogen Sulfide and Gut Microbiota: Their Synergistic Role in Modulating Sirtuin Activity and Potential Therapeutic Implications for Neurodegenerative Diseases.Pharmaceuticals (Basel). 2024 Nov 4;17(11):1480. doi: 10.3390/ph17111480. Pharmaceuticals (Basel). 2024. PMID: 39598392 Free PMC article. Review.
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
Cited by
-
The multifaceted role of sirtuins in inflammatory bowel diseases.Am J Physiol Gastrointest Liver Physiol. 2025 Jul 1;329(1):G58-G68. doi: 10.1152/ajpgi.00311.2024. Epub 2025 Apr 29. Am J Physiol Gastrointest Liver Physiol. 2025. PMID: 40298096 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
