Polycomb repressive complex 2 facilitates the transition from heterotrophy to photoautotrophy during seedling emergence
- PMID: 40515680
- PMCID: PMC12236341
- DOI: 10.1093/plcell/koaf148
Polycomb repressive complex 2 facilitates the transition from heterotrophy to photoautotrophy during seedling emergence
Abstract
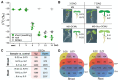
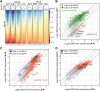
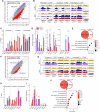
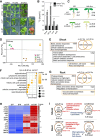
The seed-to-seedling transition represents a key developmental and metabolic switch in plants. Catabolism of seed storage reserves fuels germination and early seedling emergence until photosynthesis is established. The seed-to-seedling developmental transition is controlled by Polycomb repressive complex 2 (PRC2). However, the coordination of PRC2 activity and its contribution to transcriptional reprogramming during seedling establishment remain unknown. By analyzing H3K27me3 re-distribution and changes in gene transcription in the shoot and root tissues of heterotrophic and photoautotrophic Arabidopsis (Arabidopsis thaliana) seedlings, we reveal 2 phases of PRC2-mediated gene repression. The first phase is independent of light and photosynthesis and results in the irreversible repression of the embryo maturation program, marked by heterotrophy and reserve storage molecule biosynthesis. The second phase is associated with the repression of metabolic pathways related to germination and early seedling emergence, and H3K27me3 deposition in this phase is sensitive to photosynthesis inhibition. We show that preventing the transcription of the PRC2-repressed glyoxylate cycle gene ISOCITRATE LYASE promotes the vegetative phase transition in PRC2-depleted plants. Our findings underscore a key role of PRC2-mediated transcriptional repression in the coordinated metabolic and developmental switches that occur during seedling emergence and emphasize the close connection between metabolic and developmental identities.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures


References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

