Single-cell RNA sequencing uncovers cellular geterogeneity of granulosa cells and provides a signature for follicular development in chicken
- PMID: 40516295
- PMCID: PMC12205347
- DOI: 10.1016/j.psj.2025.105430
Single-cell RNA sequencing uncovers cellular geterogeneity of granulosa cells and provides a signature for follicular development in chicken
Abstract
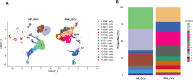
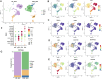
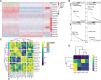
Functional differentiation of the granulosa cell (GC) layer plays an essential role in follicular development and oocyte maturation in birds. However, despite the importance of GCs, heterogeneity within the granulosa layer across poultry species remains poorly defined and functional transcriptomic signatures across distinct cellular subpopulations are lacking. In this study, single-cell RNA sequencing was used to identify GC types, uncover heterogeneity, and construct the developmental trajectories of chicken GCs at two developmental stages: the hierarchical follicle (HF)-GC and prehierarchical follicle (PHF)-GC stages. The following four GC types were identified: rapid growth, early, luteal, and primitive GCs. We found significant differences in the abundance of these different cell types at the HF-GC and PHF-GC stages. We also identified four potential differentiation trajectories for GCs during follicular development. The four distinct developmental trajectories may elucidate that the dynamic interplay and transition among these four GC types are pivotal in determining the fate of the follicle. A single-cell regulatory network inference and clustering analysis was performed to identify specifically expressed transcription factors in the different GC types. These transcription factors may have key regulatory roles in follicular development. Furthermore, we identified genes that were differentially expressed in HF-GCs and PHF-GCs. In total, 1,049 differentially expressed genes were identified in the two groups, including 379 upregulated and 670 downregulated genes. Finally, we identified several genes that may play important roles in follicle selection and development in chicken, including CDK2, CCND1, CCND2, BCL2, FOXO1, AMH, WT1, STAR, CYP11A1, HSD11B2, CYP51A1, and ESR1. These findings enhance our understanding of the cell biology of the GC layer in ovarian follicles and provide a basis for future studies of the relevant molecular regulatory mechanisms.
Keywords: Cell heterogeneity; Chicken; Granulosa cell Follicular development; Single-cell transcriptome.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



Similar articles
-
Expression and regulation of adrenomedullin 2 gene in ovarian follicles and its effect on follicular granulosa cells in chicken.Poult Sci. 2025 Jul;104(7):105204. doi: 10.1016/j.psj.2025.105204. Epub 2025 Apr 22. Poult Sci. 2025. PMID: 40300324
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Epigenetic age acceleration in follicular fluid and markers of ovarian response among women undergoing IVF.Hum Reprod. 2024 Sep 1;39(9):2003-2009. doi: 10.1093/humrep/deae136. Hum Reprod. 2024. PMID: 38890131 Free PMC article.
-
Individualised gonadotropin dose selection using markers of ovarian reserve for women undergoing in vitro fertilisation plus intracytoplasmic sperm injection (IVF/ICSI).Cochrane Database Syst Rev. 2018 Feb 1;2(2):CD012693. doi: 10.1002/14651858.CD012693.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2024 Jan 4;1:CD012693. doi: 10.1002/14651858.CD012693.pub3. PMID: 29388198 Free PMC article. Updated.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
-
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., van den Oord J., Atak Z.K., Wouters J., Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

