CRAMP1-dependent histone H1 biogenesis is essential for topoisomerase II inhibitor tolerance
- PMID: 40516529
- PMCID: PMC12240685
- DOI: 10.1016/j.molcel.2025.04.006
CRAMP1-dependent histone H1 biogenesis is essential for topoisomerase II inhibitor tolerance
Abstract
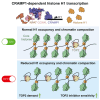
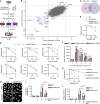
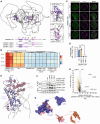
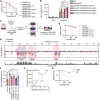
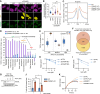
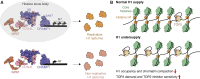
Topoisomerase II (TOP2) inhibitors (TOP2i) are mainstay chemotherapeutic agents that undermine genome integrity by stabilizing TOP2-DNA complexes accompanied by DNA damage formation. Here, we reveal the uncharacterized protein CRAMP1 and H1 linker histones as key effectors of TOP2i tolerance in human cells. We demonstrate that CRAMP1 defines a dedicated histone H1 biogenesis factor stimulating transcription of both replicative and non-replicative H1 genes, driven by its concurrent targeting to histone gene loci and H1-specific promoter motifs. CRAMP1 promotes TOP2i tolerance by maintaining H1 supply, involving a novel mechanism uncoupled from TOP2i-induced DNA damage whereby reducing the H1 pool triggers unscheduled TOP2 substrate formation in low-accessibility chromatin states. This amplifies total demand for TOP2 activity, lowering the threshold for TOP2i-mediated exhaustion of TOP2. Our discoveries elucidate the mechanistic basis of histone H1 biogenesis in human cells, opening opportunities for selectively manipulating linker but not core histone supply and targeting cancer-associated H1 deficiency.
Keywords: chromatin; genome maintenance; histone H1; histone biogenesis; histone locus bodies; histone transcription; topoisomerase II; topoisomerase II inhibitors.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

