Purine nucleoside phosphorylase dominates Influenza A virus replication and host hyperinflammation through purine salvage
- PMID: 40517177
- PMCID: PMC12167387
- DOI: 10.1038/s41392-025-02272-1
Purine nucleoside phosphorylase dominates Influenza A virus replication and host hyperinflammation through purine salvage
Abstract
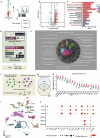
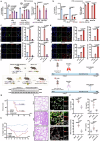
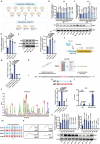
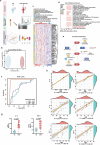
Influenza A virus (IAV) poses a significant threat to human health. The outcome of IAV results from the viral-host interaction, with the underlying molecular mechanisms largely unknown. By integrating the plasma proteomics data of the IAV-infected patients into the viral-inflammation protein-protein interaction (VI-PPI) network created in this study, purine nucleoside phosphorylase (PNP), the critical enzyme in purine salvage, was identified as a potential hub gene that connected the different stages of IAV infection. Extended survival rates and reduced pulmonary inflammatory lesions were observed in alveolar epithelial cell (AEC)-specific PNP conditional knockout mice upon H1N1 infection. Mechanistically, PB1-F2 of IAV was revealed as a novel viral transcriptional factor to bind to the TATA box of PNP promoter, leading to enhanced purine salvage in H1N1-challenged AECs. The activation of PNP-mediated purine salvage was verified in IAV-infected patients and A549 cells. PNP knockdown elicited a purine metabolic shift from augmented salvage pathway to de novo synthesis, constraining both viral infection and pro-inflammatory signaling through APRT-AICAR-AMPK activation. Moreover, durdihydroartemisinin (DHA), predicted by VI-PPI as a novel PNP inhibitor, exerted beneficial effects on the survival and weight gain of H1N1-challenged mice via its direct binding to PNP. To reveal for the first time, we found that PNP, activated by IAV, plays a hub role within H1N1-host interaction, simultaneously modulating viral replication and hyperinflammation through purine salvage. Our study sheds new light on a "two-for-one" strategy by targeting purine salvage in combating IAV-related pathology, suggesting PNP as a potential novel anti-influenza host target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Virulent PB1-F2 residues: effects on fitness of H1N1 influenza A virus in mice and changes during evolution of human influenza A viruses.Sci Rep. 2018 May 10;8(1):7474. doi: 10.1038/s41598-018-25707-y. Sci Rep. 2018. PMID: 29749408 Free PMC article.
-
H5N1 Influenza A Virus PB1-F2 Relieves HAX-1-Mediated Restriction of Avian Virus Polymerase PA in Human Lung Cells.J Virol. 2018 May 14;92(11):e00425-18. doi: 10.1128/JVI.00425-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29563290 Free PMC article.
-
Robust expression of p27Kip1 induced by viral infection is critical for antiviral innate immunity.Cell Microbiol. 2020 Nov;22(11):e13242. doi: 10.1111/cmi.13242. Epub 2020 Aug 1. Cell Microbiol. 2020. PMID: 32596986
-
Unique Transcriptional Architecture in Airway Epithelial Cells and Macrophages Shapes Distinct Responses following Influenza Virus Infection Ex Vivo.J Virol. 2019 Mar 5;93(6):e01986-18. doi: 10.1128/JVI.01986-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626665 Free PMC article.
-
Restriction of influenza A virus replication by host DCAF7-CRL4B axis.J Virol. 2025 Apr 15;99(4):e0013325. doi: 10.1128/jvi.00133-25. Epub 2025 Mar 27. J Virol. 2025. PMID: 40145735 Free PMC article.
References
-
- Gu, C. et al. A human isolate of bovine H5N1 is transmissible and lethal in animal models. Nature636, 711–718 (2024). - PubMed
-
- Uyeki, T. M. et al. Highly pathogenic avian influenza A(H5N1) virus infection in a dairy farm worker. N. Engl. J. Med.390, 2028–2029 (2024). - PubMed
-
- Gupta, N. & Silke, J. Blocking cell death limits lung damage and inflammation from influenza. Nature628, 726–727 (2024). - PubMed
-
- Leung, R. C. et al. Global emergence of neuraminidase inhibitor-resistant influenza A(H1N1)pdm09 viruses with I223V and S247N mutations: implications for antiviral resistance monitoring. Lancet Microbe5, 627–628 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

