Prospecting microbiota of Adriatic fish: Bacillus velezensis as a potential probiotic candidate
- PMID: 40517265
- PMCID: PMC12167591
- DOI: 10.1186/s42523-025-00429-5
Prospecting microbiota of Adriatic fish: Bacillus velezensis as a potential probiotic candidate
Abstract
Background: Aquaculture is one of the fastest growing sectors of food production and covers more than half of the market demand for fish and fishery products. However, aquaculture itself faces numerous challenges, such as infectious disease outbreaks, which are one of the limiting factors for the growth and environmental sustainability of modern aquaculture. Understanding the composition and diversity of the gut microbiota of fish is important to elucidate its role in host health and aquaculture management. In addition, the gut microbiota represents a valuable source of bacteria with probiotic potential for farmed fish.
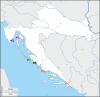
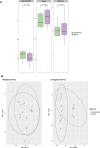
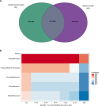
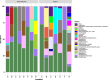
Results: In this study, we analysed the intestinal microbiota of two economically important fish species, the European seabass (Dicentrarchus labrax) and the gilthead seabream (Sparus aurata), using 16S rRNA gene amplicon sequencing. The taxonomic analysis identified 462 amplicon sequence variants at a similarity level of 99 and showed similar alpha diversity indices between seabass and gilthead seabream. Beta diversity analysis showed no significant differentiation in gut microbiota between fish species or aquaculture sites. Among the culturable isolates, a high proportion of Photobacterium damselae and Bacillus spp. was detected. We selected a single Bacillus velezensis isolate and further characterised its biosynthetic potential by performing whole genome sequencing. Its genome contains biosynthetic gene clusters for most of the common secondary metabolites typical of B. velezensis. Antibiotic susceptibility testing showed the sensitivity of the selected isolates to several antibiotics according to EFSA recommendations. Furthermore, stimulation of peripheral blood leukocytes (PBL) with B. velezensis resulted in a strong pro-inflammatory response, with a pronounced upregulation of cytokines il1b, il6, tnfa and il10 observed over time.
Conclusions: Overall, this study provides an insight into the composition of the intestinal microbiota and the diversity of culturable intestinal bacteria of two economically most important fish species from Adriatic cage culture and sheds light on the autochthonous intestinal B. velezensis as a promising probiotic candidate for Mediterranean aquaculture.
Keywords: Bacillus velezensis; Dicentrarchus labrax; Sparus aurata; Aquaculture; Probiotics; Whole genome sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- FAO. In brief to the state of world fisheries and aqauculture 2024. Blue Transformation in Action. Nat. Resour. Rome; 2024.
-
- FEAP. European Aquaculture Production Report 2016–20222. FEAP Prod. Rep. 2023. Available from: https://feap.info/wp-content/uploads/2020/10/20201007_feap-production-re...
-
- Hellenic Aquaculture Producers Organisation (HAPO). Aquaculture in Greece - Annual report 2023. 2023;1–52. Available from: https://fishfromgreece.com/wp-content/uploads/2023/10/HAPO_AR23_WEB-NEW.pdf
-
- Maldonado-Miranda JJ, Castillo-Pérez LJ, Ponce-Hernández A, Carranza-Álvarez C. Summary of economic losses due to bacterial pathogens in aquaculture industry. In: Dar HG, Bhat RA, Qadri H, Al-Ghamdy KM, Hakeem KR, editors. Bact Fish Dis. Academic Press; 2022. p. 399–417.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
