Staphylococci in high resolution: Capturing diversity within the human nasal microbiota
- PMID: 40517386
- PMCID: PMC12322312
- DOI: 10.1016/j.celrep.2025.115854
Staphylococci in high resolution: Capturing diversity within the human nasal microbiota
Abstract
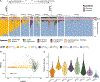
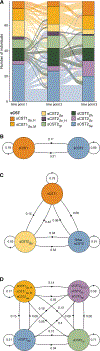
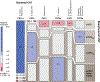
Staphylococci include both nasal commensals and opportunistic pathogens, globally responsible for a large proportion of infection-related deaths, especially in S. aureus carriers. To understand staphylococcal temporal dynamics within the nasal microbiota, we employed Staphylococcus-targeted sequencing in two cohorts from Denmark and Germany. We identified two major staphylococcal community state types (sCSTs)-one dominated by S. aureus and one dominated by S. epidermidis-and eight subgroups defined by co-colonizing coagulase-negative staphylococci. The distribution of sCSTs was similar between the two cohorts. Predominance of either S. aureus or S. epidermidis was highly persistent over time, whereas co-colonizing staphylococcal species were transient with varying stability among the sCST subgroups. Detection of S. aureus by culture was positively associated with absolute abundance by qPCR. S. aureus domination was diminished when Dolosigranulum and Corynebacterium co-occurred. Our findings could inform efforts to reduce S. aureus nasal colonization and infection.
Keywords: CP: Microbiology; Staphylococcus aureus; Staphylococcus epidermidis; antagonist; carriage; community state type; microbial abundance; microbiome; nasal microbiota; staphylococci.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.M.L. is a founder of and holds equity in Trench Therapeutics. L.B.P. has received research support from Trench Therapeutics.
Figures

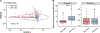


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

