Comprehensive analysis of Enterococcus spp. from two European healthy infant cohorts shows stable genomic traits including antimicrobial resistance (AMR)
- PMID: 40518985
- PMCID: PMC12184193
- DOI: 10.1080/19490976.2025.2516699
Comprehensive analysis of Enterococcus spp. from two European healthy infant cohorts shows stable genomic traits including antimicrobial resistance (AMR)
Abstract
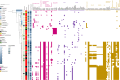
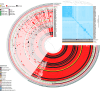
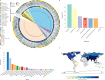
Enterococcus spp. some of which are pathogenic, are common gut microbiota members, including also infants. Infants may be more susceptible to Enterococcus due to their developing gut ecosystems. It is unclear whether antibiotic resistance genes (ARGs) and certain genomic traits in enterococci are restricted to the human subpopulation or more widespread. Furthermore, the correlation between these traits and geographic variation is poorly understood. Therefore, we sequenced 100 strains isolated from full-term healthy infants' fecal samples from two geographically distant European cohorts (MAMI in Spain and LucKi from the Netherlands) to explore the diversity of Enterococcus spp. within the infant's gut microbiome and assess cohort-specific traits such as ARGs. Most isolates were E. faecalis and E. gallinarum, with a total of 11 species identified. We found a rich reservoir of ARGs, plasmids, prophages and virulence factors in the infant strains, with minimal cohort-specific differences in resistome profiles. In addition, Epx, a pore-forming toxin associated with pathogenicity, was found in E. hirae strains. While metabolic profiles were similar across cohorts, E. faecalis strains harbored more virulence genes and prophages compared to other species. An analysis of public Enterococcus genomes revealed that multi-drug resistant (MDR) strains exist without any significant geographic or temporal pattern. Phenotypic resistance analysis indicated that 28% of MAMI strains were gentamicin resistant, compared to 5% of the strains from the LucKi cohort, though LucKi isolates were also resistant to other antibiotics. We also selected ten E. faecalis isolates with varying virulence gene repertoires for phenotypic virulence testing in Caenorhabditis elegans and found them killing at various rates, however no clear pattern emerged in correlation with any specific genetic determinant. Overall, our results suggest that Enterococcus spp. including ARGs, are highly mobile across Europe and beyond. Their adaptability likely facilitates long-distance dissemination, with strains being acquired early in life from community environments.
Keywords: AMR; Enterococcus; genomics; healthy infants; phylogenomics; resistome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Boehm AB, Sassoubre LM. Enterococci as indicators of environmental fecal contamination. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Massachusetts Eye and Ear Infirmary. Boston; 2014. https://www.ncbi.nlm.nih.gov/books/NBK190421/ - PubMed
-
- Neuhaus K, Lamparter MC, Zölch B, Landstorfer R, Simon S, Spanier B, Ehrmann MA, Vogel RF. Probiotic enterococcus faecalis Symbioflor® down regulates virulence genes of EHEC in vitro and decrease pathogenicity in a Caenorhabditis elegans model. Arch Microbiol. 2017;199(2):203–23. doi: 10.1007/s00203-016-1291-8. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
