Transcriptome and metabolome profiling reveal the chlorogenic acid as a resistance substance for rice against the white-backed planthopper Sogatella furcifera (Horváth)
- PMID: 40519591
- PMCID: PMC12162631
- DOI: 10.3389/fpls.2025.1571893
Transcriptome and metabolome profiling reveal the chlorogenic acid as a resistance substance for rice against the white-backed planthopper Sogatella furcifera (Horváth)
Abstract
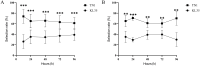
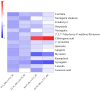
The white-backed planthopper (WBPH), Sogatella furcifera (Horváth) is a major migratory pest of rice, making research on rice resistance to WBPH essential for rice breeding and pest management. This study compared the resistance of susceptible rice TN1 and resistant rice KL35 to WBPH by analyzing antixenosis, antibiosis, and tolerance. We also conducted transcriptome and metabolome analysis to identify the defensive compounds against the WBPH and regulatory genes in KL35. The results indicated that KL35 exhibited significant antixenosis and tolerance to WBPH, markedly prolonging developmental duration and reducing fecundity. Metabolomic analysis identified 15 core metabolites, among which chlorogenic acid (CGA) content in KL35 was significantly higher than in TN1 both before and after WBPH feeding. Integrated transcriptomic and metabolomic analyses showed that the flavonoid biosynthetic pathway was a key anti-pest pathway in KL35. Additionally, two genes cinnamate 4-hydroxylase gene (Os05g0320700) and 4-coumarate CoA ligase (Os02g0697400) were identified and postulated as key players in the CGA biosynthesis pathway in KL35. Exogenous application of CGA to TN1 enhanced its tolerance and antixenosis to WBPH, significantly decreasing WBPH's survival and mean dry weight. These findings suggest that CGA is an important resistance substance against WBPH. As a plant-derived and environment-friendly compound, CGA could be a potentially important compound for rice WBPH resistance agriculture.
Keywords: chlorogenic acid; metabolome; resistance; rice; transcriptome; white-backed planthopper.
Copyright © 2025 Xie, Tao, Zhang, Luo, Deng, Li, Li, Wang, Yue, Jiang and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Asif S., Jang Y. H., Kim E. G., Jan R., Asaf S., Aaqil Khan M., et al. (2022). The role of exogenous gibberellic acid and methyl jasmonate against white-backed planthopper (Sogatella furcifera) stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 23, 14737. doi: 10.3390/ijms232314737 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

