Sterols in plant biology - Advances in studying membrane dynamics
- PMID: 40519718
- PMCID: PMC12167035
- DOI: 10.1016/j.tcsw.2025.100147
Sterols in plant biology - Advances in studying membrane dynamics
Abstract
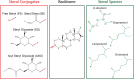
Plants sense their environment at the cell surface, i.e. the plasma membrane, where extracellular signals are perceived and transduced. Together with the cortical cytoskeleton and the cell wall, membrane lipids can influence these processes by acting on protein dynamics at the plasma membrane. Among these lipids, sterols regulate membrane fluidity and thus, protein functions. However, plant sterols are diverse in structure and particularly difficult to study due to technical limitations. Nevertheless, advances in sterol imaging, sterol-protein interaction studies, and sterol perturbation methods have resulted in a better understanding of their functions in plant development and physiology. Here we summarize the current knowledge and the latest breakthroughs, and discuss future challenges, in the field of plant sterol biology and cell surface organization.
Keywords: Lipid-order; Membrane-contact sites; Phytosterols; Plant membrane biology; Sterol conjugates; Sterol dynamics; Sterol transport; Sterols.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Plant phloem sterol content: forms, putative functions, and implications for phloem-feeding insects.Front Plant Sci. 2013 Sep 24;4:370. doi: 10.3389/fpls.2013.00370. eCollection 2013. Front Plant Sci. 2013. PMID: 24069026 Free PMC article.
-
Differential effect of plant lipids on membrane organization: specificities of phytosphingolipids and phytosterols.J Biol Chem. 2015 Feb 27;290(9):5810-25. doi: 10.1074/jbc.M114.598805. Epub 2015 Jan 9. J Biol Chem. 2015. PMID: 25575593 Free PMC article.
-
Sterol dynamics during endocytic trafficking in Arabidopsis.Methods Mol Biol. 2014;1209:13-29. doi: 10.1007/978-1-4939-1420-3_2. Methods Mol Biol. 2014. PMID: 25117272
-
Plant Sterols: Diversity, Biosynthesis, and Physiological Functions.Biochemistry (Mosc). 2016 Aug;81(8):819-34. doi: 10.1134/S0006297916080046. Biochemistry (Mosc). 2016. PMID: 27677551 Review.
-
Noncholesterol sterols.Acta Univ Carol Med Monogr. 2008;154:5-101. Acta Univ Carol Med Monogr. 2008. PMID: 19283968 Review.
Cited by
-
The Evolution of Plant Hormones: From Metabolic Byproducts to Regulatory Hubs.Int J Mol Sci. 2025 Jul 25;26(15):7190. doi: 10.3390/ijms26157190. Int J Mol Sci. 2025. PMID: 40806323 Free PMC article. Review.
References
-
- Asami T., Masaharu Mizutani, Yukihisa Shimada, Hideki Goda, Nobutaka Kitahata, Katsuhiko Sekimata, Han S.-Y., et al. Triadimefon, a Fungicidal Triazole-Type P450 Inhibitor, Induces Brassinosteroid Deficiency-like Phenotypes in Plants and Binds to DWF4 Protein in the Brassinosteroid Biosynthesis Pathway. Biochemical Journal. 2003;369(Pt 1):71–76. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

