Proteome-wide characterization of PTMs reveals host cell responses to viral infection and identifies putative antiviral drug targets
- PMID: 40519923
- PMCID: PMC12162599
- DOI: 10.3389/fimmu.2025.1587106
Proteome-wide characterization of PTMs reveals host cell responses to viral infection and identifies putative antiviral drug targets
Erratum in
-
Correction: Proteome-wide characterization of PTMs reveals host cell responses to viral infection and identifies putative antiviral drug targets.Front Immunol. 2025 Aug 12;16:1672766. doi: 10.3389/fimmu.2025.1672766. eCollection 2025. Front Immunol. 2025. PMID: 40873564 Free PMC article.
Abstract
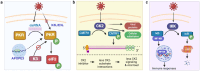
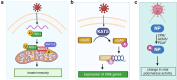
Post-translational modifications (PTMs) are biochemical modifications that can significantly alter protein structure, function, stability, localization, and interactions with other molecules, thereby activating or inactivating intracellular processes. A growing body of research has begun to highlight the role of PTMs, including phosphorylation, ubiquitination, acetylation, and redox modifications, during virus-host interactions. Collectively, these PTMs regulate key steps in mounting the host immune response and control critical host pathways required for productive viral replication. This has led to the conception of antiviral therapeutics that focus on controlling host protein PTMs, potentially offering pathogen-agnostic treatment options and revolutionizing our capacity to prevent virus transmission. On the other hand, viruses can hijack the host cellular PTM machinery to modify viral proteins in promoting viral replication and evading immune surveillance. PTM regulation during virus-host interactions is complex and poorly mapped, and the development of effective PTM-targeted antiviral drugs will require a more comprehensive understanding of the cellular pathways essential for virus replication. In this review, we discuss the roles of PTMs in virus infection and how technological advances in mass spectrometry-based proteomics can capture systems-level PTM changes during viral infection. Additionally, we explore how such knowledge is leveraged to identify PTM-targeted candidates for developing antiviral drugs. Looking ahead, studies focusing on the discovery and functional elucidation of PTMs, either on the host or viral proteins, will not only deepen our understanding of molecular pathology but also pave the way for developing better drugs to fight emerging viruses.
Keywords: PTMs; acetylation; antiviral drug; phosphorylation; proteome; redox; ubiquitination; viral infection.
Copyright © 2025 Li, Kabza, Ives, Thiel, Waters, Qian, Sims and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

