Emodin Inhibits NLRP3 Inflammasome Activation and Protects Against Sepsis via Promoting FUNDC1-Mediated Mitophagy
- PMID: 40520006
- PMCID: PMC12160860
- DOI: 10.7150/ijbs.110904
Emodin Inhibits NLRP3 Inflammasome Activation and Protects Against Sepsis via Promoting FUNDC1-Mediated Mitophagy
Abstract
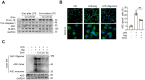
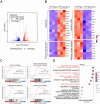
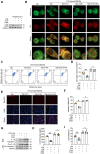
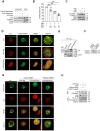
Dysregulated activation of the NLR family pyrin domain-containing 3 (NLRP3) inflammasome contributes to the pathogenesis of numerous inflammatory and infectious diseases; however, effective targeted therapies remain elusive. In this study, we identify emodin-a bioactive anthraquinone derived from Rheum palmatum (radix Rhei) and Polygonum cuspidatum (Polygonaceae)-as a potent and selective inhibitor of NLRP3 inflammasome activation. Notably, emodin disrupts the assembly of the NLRP3 complex without impairing inflammasome priming. Transcriptomic profiling via RNA sequencing reveals that emodin reprograms mitochondrial quality control pathways, markedly enhancing mitophagy flux. Mechanistically, emodin suppresses casein kinase II (CK2)-mediated phosphorylation of FUNDC1, a pivotal mitophagy receptor, thereby promoting mitochondrial clearance and preventing mitochondrial reactive oxygen species-induced NLRP3 inflammasome assembly. Both genetic silencing of FUNDC1 and pharmacological inhibition of mitophagy with 3-methyladenine abrogated abrogate the inhibitory effects of emodin, establishing a direct mechanistic link between FUNDC1-dependent mitophagy and NLRP3 regulation. In vivo, emodin confers significant protection in sepsis models, with these protective effects being lost in NLRP3-deficient mice or upon macrophage-specific deletion of FUNDC1. Collectively, our findings uncover a novel CK2-FUNDC1-mitophagy axis through which emodin inhibits NLRP3 inflammasome activation, highlighting its promise as a clinically translatable candidate for the treatment of NLRP3-driven inflammatory diseases.
Keywords: Emodin; FUNDC1; Mitochondrial homeostasis; Mitophagy; NLRP3 inflammasome; Sepsis..
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Xing Y, Yao X, Li H, Xue G, Guo Q, Yang G. et al. Cutting Edge: TRAF6 Mediates TLR/IL-1R Signaling-Induced Nontranscriptional Priming of the NLRP3 Inflammasome. J Immunol. 2017;199:1561–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

