Luteolin improves precancerous conditions of the gastric mucosa by binding STAT3 and inhibiting LCN2 expression
- PMID: 40520011
- PMCID: PMC12160510
- DOI: 10.7150/ijbs.111636
Luteolin improves precancerous conditions of the gastric mucosa by binding STAT3 and inhibiting LCN2 expression
Abstract
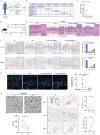
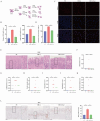
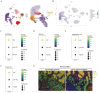
Inhibition of malignant transformation from the precancerous stage has important clinical value for the prevention of gastric cancer. Here, we report a strategy to inhibit precancerous gastric conditions by Luteolin (Lut). Lut treatment resulted in remarkable resistance to oxyntic atrophy, spasmolytic polypeptide-expressing metaplasia (SPEM), and gastric mucosal injury in tamoxifen (TAM)-treated mice, chenodeoxycholic acid-treated rats, and human organoids. Mechanism study suggested that LCN2 expression was upregulated in the SPEM mucosa and downregulated after Lut treatment. LCN2 blocking suppressed TAM-induced oxyntic atrophy and metaplasia and partially counteracted the effect of Lut. Quantitative chemoproteomics identified that Lut bound to STAT3 and inhibited its phosphorylation. Functional experiments using STAT3 inhibitors and epithelial cell-specific Stat3 deficient mice showed that STAT3 inhibition and deletion attenuated the beneficial effects of Lut. Our data supported that Lut might be a therapeutic candidate for the treatment of gastric mucosal injury by binding to STAT3 and thereby inhibiting the STAT3/LCN2 axis.
Keywords: Gastric cancer; LCN2; Luteolin; Precancerous gastric conditions; STAT3; Spasmolytic polypeptide-expressing metaplasia.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

