Radio-chemotherapy and metformin selectively modulate the heterogeneous landscape of glioma with ribosome biogenesis, long non coding RNA and immune-escape markers as major player
- PMID: 40520013
- PMCID: PMC12160513
- DOI: 10.7150/ijbs.103194
Radio-chemotherapy and metformin selectively modulate the heterogeneous landscape of glioma with ribosome biogenesis, long non coding RNA and immune-escape markers as major player
Abstract
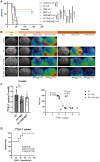
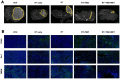
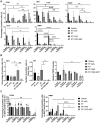
Glioblastoma multiforme (GBM) is the most common primary brain tumor in adults with a short survival time after standard therapy administration including radiotherapy (RT) associated with temozolomide (TMZ). Here, we investigated the effects of radiochemotherapy in association with metformin (MET), a drug targeting cell metabolism on a syngeneic GBM mouse model using Positron Emission Tomography imaging with [18F]FLT and [18F]VC701 and single-cell RNA-sequencing analysis. The addition of drugs to RT significantly increased survival and [18F]FLT showed an early predictive response of combined therapy. We identified the presence of heterogeneous tumor populations with different treatment sensitivity and a complex immune evasive microenvironment. Tumor cells surviving to treatments showed immune response, among the main differentially modulated biological functions and a potential role of long non-coding RNAs (lncRNAs) in treatment resistance. Association with TMZ or TMZ plus MET reduced the pro-tumor phenotype of immune reaction acting more on myeloid cells the first and on lymphocytes the latter. Off note, MET add-on counteracted the immune-evasive phenotype particularly of T cells suggesting a potential role of MET also in adopted immunity.
Keywords: Glioblastoma; LAG3; PET imaging; long-non coding RNA; metformin; radio-chemotherapy; single-cell RNA sequencing.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures









References
-
- Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJB. et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005 Mar 10;352(10):987–96. - PubMed
-
- Rabab'h O, Al-Ramadan A, Shah J, Lopez-Negrete H, Gharaibeh A. Twenty Years After Glioblastoma Multiforme Diagnosis: A Case of Long-Term Survival. Cureus [Internet]. 2021 Jun 30 [cited. 2024. May 21]; Available from: https://www.cureus.com/articles/60104-twenty-years-after-glioblastoma-mu.... - PMC - PubMed
-
- Zhang CS, Li M, Ma T, Zong Y, Cui J, Feng JW. et al. Metformin Activates AMPK through the Lysosomal Pathway. Cell Metab. 2016 Oct;24(4):521–2. - PubMed
-
- Wu X yu, Xu WW, Huan X kun, Wu G nan, Li G, Zhou YH. et al. Mechanisms of cancer cell killing by metformin: a review on different cell death pathways. Mol Cell Biochem. 2023 Jan;478(1):197–214. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

