The chromosomal genome sequence of the sponge Crambe crambe (Schmidt, 1862) and its associated microbial metagenome sequences
- PMID: 40520149
- PMCID: PMC12166358
- DOI: 10.12688/wellcomeopenres.24154.1
The chromosomal genome sequence of the sponge Crambe crambe (Schmidt, 1862) and its associated microbial metagenome sequences
Abstract
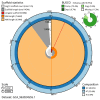
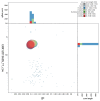
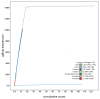
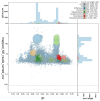
We present a genome assembly from an individual Crambe crambe (Porifera; Demospongiae; Poecilosclerida; Crambeidae). The host genome sequence is 143.20 megabases in span. Most of the assembly is scaffolded into 18 chromosomal pseudomolecules. The mitochondrial genome has also been assembled and is 19.53 kilobases in length. Several symbiotic prokaryotic genomes were assembled as MAGs, including two relevant sponge symbionts, the Candidatus Beroebacter blanensis/ AqS2 clade (Tethybacterales, Gammaproteobacteria) of LMA sponges, and the widely distributed archaeal Nitrosopumilus sp. clade.
Keywords: Crambe crambe; Poecilosclerida; chromosomal; genome sequence; marine sponge.
Copyright: © 2025 Maldonado M et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Becerro MA, Uriz MJ, Turón X: Trends in space occupation by the encrusting sponge Crambe crambe: variation in shape as a function of size and environment. Mar Biol. 1994;121:301–307. 10.1007/BF00346738 - DOI
LinkOut - more resources
Full Text Sources

