Harnessing dual variational autoencoders to decode microbe roles in diseases for traditional medicine discovery
- PMID: 40520163
- PMCID: PMC12162629
- DOI: 10.3389/fphar.2025.1578140
Harnessing dual variational autoencoders to decode microbe roles in diseases for traditional medicine discovery
Abstract
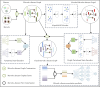
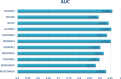
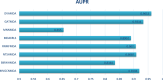
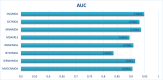
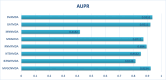
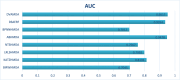
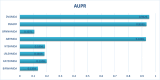
Traditional medicine encompasses a rich trove of knowledge and practices for disease prevention, diagnosis, and treatment. However, it faces challenges such as poorly defined compositions of preparations and limited high-quality efficacy data. The development of artificial intelligence presents new opportunities for traditional medicine research and applications, especially in predicting MDAs (MDAs), which is of great significance for understanding disease mechanisms and developing new treatments. This study proposes a MDAs prediction method based on double variational autoencoders (DVAMDA). This method innovatively integrates double variational autoencoders and multi-information fusion techniques. Firstly, the graph SAGE encoder is utilized to preliminarily extract the local and global structural information of nodes. Subsequently, the double variational autoencoders are employed to separately extract the latent probability distribution information of the initial input data and the graph-specific property information from the output of the graph SAGE encoder. Then, these different sources of information are fused to provide rich and powerful feature support for subsequent prediction tasks. Finally, the Hadamard product operation and a deep neural network are used to predict MDAs. Experimental results on the HMDAD and Disbiome datasets show that the DVAMDA model performs outstandingly in multiple evaluation metrics. The findings of this research contribute to a deeper understanding of microbe-disease relationships and provide strong support for drug development in traditional medicine based on MDAs. The relevant data and code are publicly accessible at: https://github.com/yxsun25/DVAMDA.
Keywords: double variational autoencoders; microbe-disease association; multi-information fusion; prediction model; traditional medicine discovery.
Copyright © 2025 Ye and Sun.
Conflict of interest statement
Author YS was employed by Zhejiang Aerospace Hengjia Data Technology Co., Ltd. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Adamic L. A., Adar E. (2003). Friends and neighbors on the Web. Social Networks, 25 (3), 211–230. 10.1016/S0378-8733(03)00009-1 - DOI
-
- Cheng E., Zhao J., Wang H., Song S., Xiong S., Sun Y. (2023). Dual network contrastive learning for predicting microbe -disease associations. IEEE/ACM Trans. Comput. Biol. Bioinforma. 20 (6), 3469–3481. 10.1109/TCBB.2022.3228617 - DOI
LinkOut - more resources
Full Text Sources

