Network-based analysis and experimental validation of identified natural compounds from Yinchen Wuling San for acute myeloid leukemia
- PMID: 40520175
- PMCID: PMC12162526
- DOI: 10.3389/fphar.2025.1591164
Network-based analysis and experimental validation of identified natural compounds from Yinchen Wuling San for acute myeloid leukemia
Abstract
Objective: Traditional Chinese medicine (TCM) has garnered attention for its potential in cancer therapy. Yinchen Wuling San (YWLS), a classical herbal formula, has been traditionally used for liver-related conditions, but its bioactive components and molecular mechanisms relevant to hematologic malignancies such as acute myeloid leukemia (AML) remain unclear. This study aims to identify the active compounds and potential molecular targets of Yinchen Wuling San in the context of AML through network pharmacology analysis, and to experimentally validate the effects of selected candidate compounds in AML models.
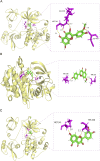
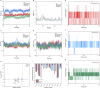
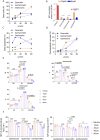
Methods: Active ingredients from six YWLS herbs were screened via the TCMSP database using oral bioavailability ≥30% and DL ≥0.18 thresholds. Targets were predicted using SwissTargetPrediction, and AML-related genes were obtained from DisGeNET and GeneCards. Key overlapping targets were analyzed via STRING PPI networks and GO/KEGG enrichment. Molecular docking was performed between three core compounds (genkwanin, isorhamnetin, quercetin) and hub proteins (e.g., SRC) using Sybyl-X. ADME profiles were predicted using SwissADME, and molecular dynamics simulations (GROMACS) assessed complex stability. These compounds were further evaluated in vitro (viability, apoptosis, cell cycle, RT-qPCR, flow cytometry) and in vivo using an AML xenograft mouse model.
Results: Of 621 YWLS targets, 113 overlapped with 1,247 AML-related genes. PPI analysis identified hub genes, including AKT1, SRC, and EGFR. Enrichment analysis highlighted PI3K-AKT, MAPK, and JAK-STAT pathways. Genkwanin, isorhamnetin, and quercetin were predicted to target SRC, with strong molecular docking affinities. ADME analysis suggested favorable pharmacokinetics, and molecular dynamics simulations confirmed structural stability. In vitro, these compounds exhibited dose-dependent cytotoxicity, induced apoptosis, modulated the cell cycle, and downregulated SRC expression. Notably, Genkwanin promoted CD8+ T cell proliferation and inhibited leukemia growth, improving survival in a leukemia xenograft model.
Conclusion: YWLS compounds, particularly Genkwanin, exhibit significant anti-leukemic activity via apoptosis induction, cell cycle modulation, and promote T cells proliferation. Genkwanin emerges as a promising therapeutic candidate for AML, warranting further clinical investigation.
Keywords: Yinchen Wuling San; acute myeloid leukemia; molecular docking; molecular dynamics simulation; network pharmacology; pharmacological target.
Copyright © 2025 Zhang, Fu, Wang and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Bertacchini J., Guida M., Accordi B., Mediani L., Martelli A. M., Barozzi P., et al. (2014). Feedbacks and adaptive capabilities of the PI3K/Akt/mTOR axis in acute myeloid leukemia revealed by pathway selective inhibition and phosphoproteome analysis. Leukemia 28 (11), 2197–2205. 10.1038/leu.2014.123 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

