Synthetic4Health: generating annotated synthetic clinical letters
- PMID: 40520216
- PMCID: PMC12163008
- DOI: 10.3389/fdgth.2025.1497130
Synthetic4Health: generating annotated synthetic clinical letters
Abstract
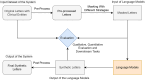
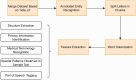
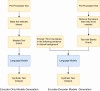
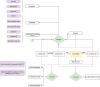
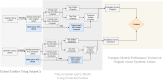
Clinical letters contain sensitive information, limiting their use in model training, medical research, and education. This study aims to generate reliable, diverse, and de-identified synthetic clinical letters to support these tasks. We investigated multiple pre-trained language models for text masking and generation, focusing on Bio_ClinicalBERT, and applied different masking strategies. Evaluation included qualitative and quantitative assessments, downstream named entity recognition (NER) tasks, and clinically focused evaluations using BioGPT and GPT-3.5-turbo. The experiments show: (1) encoder-only models perform better than encoder-decoder models; (2) models trained on general corpora perform comparably to clinical-domain models if clinical entities are preserved; (3) preserving clinical entities and document structure aligns with the task objectives; (4) Masking strategies have a noticeable impact on the quality of synthetic clinical letters: masking stopwords has a positive impact, while masking nouns or verbs has a negative effect; (5) The BERTScore should be the primary quantitative evaluation metric, with other metrics serving as supplementary references; (6) Contextual information has only a limited effect on the models' understanding, suggesting that synthetic letters can effectively substitute real ones in downstream NER tasks; (7) Although the model occasionally generates hallucinated content, it appears to have little effect on overall clinical performance. Unlike previous research, which primarily focuses on reconstructing original letters by training language models, this paper provides a foundational framework for generating diverse, de-identified clinical letters. It offers a direction for utilizing the model to process real-world clinical letters, thereby helping to expand datasets in the clinical domain. Our codes and trained models are available at https://github.com/HECTA-UoM/Synthetic4Health.
Keywords: clinical NLP (natural language processing); encoder-only models; encoder–decoder models; masking and generating; named entity recognation; pre-trained language models (PLMs); synthetic data creation.
© 2025 Ren, Belkadi, Han, Del-Pinto and Nenadic.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









Similar articles
-
Using Synthetic Health Care Data to Leverage Large Language Models for Named Entity Recognition: Development and Validation Study.J Med Internet Res. 2025 Mar 18;27:e66279. doi: 10.2196/66279. J Med Internet Res. 2025. PMID: 40101227 Free PMC article.
-
Are synthetic clinical notes useful for real natural language processing tasks: A case study on clinical entity recognition.J Am Med Inform Assoc. 2021 Sep 18;28(10):2193-2201. doi: 10.1093/jamia/ocab112. J Am Med Inform Assoc. 2021. PMID: 34272955 Free PMC article.
-
DeIDNER Corpus: Annotation of Clinical Discharge Summary Notes for Named Entity Recognition Using BRAT Tool.Stud Health Technol Inform. 2021 May 27;281:432-436. doi: 10.3233/SHTI210195. Stud Health Technol Inform. 2021. PMID: 34042780 Free PMC article.
-
Pre-trained language models in medicine: A survey.Artif Intell Med. 2024 Aug;154:102904. doi: 10.1016/j.artmed.2024.102904. Epub 2024 Jun 5. Artif Intell Med. 2024. PMID: 38917600 Review.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
References
LinkOut - more resources
Full Text Sources

