Flexible iron: disorder in the ironome brings order to protein structure and function
- PMID: 40520260
- PMCID: PMC12162314
- DOI: 10.3389/fmolb.2025.1537164
Flexible iron: disorder in the ironome brings order to protein structure and function
Abstract
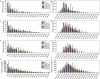
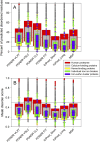
Iron is one of the most abundant elements on earth. The most recognized role of iron in living organisms is its incorporation in the heme-containing protein hemoglobin, which is abundantly found in the red blood cells that facilitate the oxygen transportation throughout the body. In fact, about 70% of organism's iron is found in hemoglobin. However, besides being essential for oxygen transport and serving as a crucial component of the molecular oxygen-carrying proteins hemoglobin and myoglobin, iron has a wide range of other biological functions. It is involved in numerous metabolic and regulatory processes and therefore is indispensable for almost all living organisms. Since iron enzymes are responsible for most of the redox metallo-catalysts, it is not surprising that 6.5% of all human enzymes are expected to be iron-dependent. Furthermore, iron-binding proteins account for about 2% of the entire proteome. The ironome encompasses heme-binding proteins, proteins binding individual iron ions, and iron-sulfur cluster-binding proteins. Although the structure-function relations of ordered iron-binding proteins are rather well understood, the prevalence and functionality of intrinsic disorder in iron-binding proteins remain to be evaluated. To fill this knowledge gap, in this study, we evaluate the intrinsic disorder of the human ironome. Our analysis revealed that the human ironome contains a noticeable level of functional intrinsic disorder, with most noticeable applications in protein-protein interactions, posttranslational modifications, and liquid-liquid phase separation.
Keywords: intrinsically disordered proteins; iron; iron-binding proteins; iron-sulfur center; liquid-liquid phase transition; proteinprotein interactions.
Copyright © 2025 Uversky and Ferreira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









Similar articles
-
Functional unfoldomics: Roles of intrinsic disorder in protein (multi)functionality.Adv Protein Chem Struct Biol. 2024;138:179-210. doi: 10.1016/bs.apcsb.2023.11.001. Epub 2023 Nov 22. Adv Protein Chem Struct Biol. 2024. PMID: 38220424 Review.
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Abundance and functional roles of intrinsic disorder in the antimicrobial peptides of the NK-lysin family.J Biomol Struct Dyn. 2017 Mar;35(4):836-856. doi: 10.1080/07391102.2016.1164077. Epub 2016 Apr 4. J Biomol Struct Dyn. 2017. PMID: 26957115
-
Intrinsic disorder-based protein interactions and their modulators.Curr Pharm Des. 2013;19(23):4191-213. doi: 10.2174/1381612811319230005. Curr Pharm Des. 2013. PMID: 23170892 Review.
References
-
- Albagli O., Dhordain P., Deweindt C., Lecocq G., Leprince D. (1995). The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 6, 1193–1198. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

