Rapid screening and taste mechanism of novel umami peptides from natural tripeptide database
- PMID: 40520701
- PMCID: PMC12166769
- DOI: 10.1016/j.fochx.2025.102565
Rapid screening and taste mechanism of novel umami peptides from natural tripeptide database
Abstract
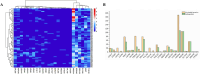
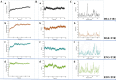
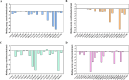
Small peptides, particularly tripeptides, play a crucial role in food umami taste. To dig for more umami tripeptides, the novel tripeptides pharmacophore model was established to rapidly screen umami peptides from natural tripeptide database. Twenty peptides with potential umami characteristics from 8000 tripeptides were further screened by molecular docking. The electronic tongue analysis and sensory evaluation suggested that the 20 tripeptides exhibited umami taste characteristics. The thresholds of the 20 tripeptides spanned from 0.137 to 2.237 mmol/L. Molecular dynamics simulations were used on T1R1 and four tripeptides with high umami intensity to reveal their taste mechanisms. In this study, a new screening strategy was established and 20 new umami tripeptides were identified and validated, providing a theoretical reference for rapid screening of umami tripeptides.
Keywords: Common characteristic pharmacophore; Molecular docking; Molecular dynamics simulation; Rapid virtual screening; Umami tripeptides.
© 2025 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- An J., Wicaksana F., Woo M.W., Liu C., Tian J., Yao Y. Current food processing methods for obtaining umami peptides from protein-rich foods: A review. Trends in Food Science & Technology. 2024;153 doi: 10.1016/j.tifs.2024.104704. - DOI
LinkOut - more resources
Full Text Sources

