Screening and Evaluation of tRF-Glu-CTC-013 as a Biomarker and Key Regulator in the Development of Cardiac Hypertrophy
- PMID: 40520901
- PMCID: PMC12163609
- DOI: 10.7150/ijms.106114
Screening and Evaluation of tRF-Glu-CTC-013 as a Biomarker and Key Regulator in the Development of Cardiac Hypertrophy
Abstract
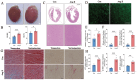
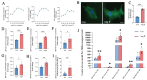
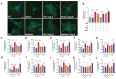
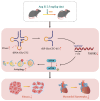
tRNA-derived small RNAs (tsRNAs) are a newly recognized class of non-coding RNAs involved in regulating RNA processing and translational control. Pathological cardiac hypertrophy, characterized by left ventricular remodeling under chronic stress, serves as a critical precursor to severe cardiovascular pathologies including myocardial ischemia, infarction, and heart failure. Utilizing an angiotensin II (Ang II)-induced mouse cardiac hypertrophy model combined with tsRNA transcriptome profiling, we identified differentially expressed tsRNAs and investigated their functional relevance. Validation in neonatal mouse ventricular myocytes (NMVMs) revealed five upregulated tsRNAs associated with hypertrophic progression. Functional characterization showed that overexpressing tRF-Glu-CTC-013 significantly reduced cardiomyocyte hypertrophy and inhibited inflammation and fibrosis. Further luciferase reporter assays revealed that tRF-Glu-CTC-013 could bind to the 3' UTR of TAS1R3, thereby inhibiting its expression and enhancing the level of autophagy in NMVMs. Taken together, these findings suggest that tsRNAs may act as novel regulators of cardiac remodeling, with tRF-Glu-CTC-013 emerging as a promising therapeutic candidate for cardioprotection via anti-hypertrophic, anti-inflammatory, and anti-fibrotic mechanisms.
Keywords: cardiac hypertrophy; tRNA-derived fragment (tRF); tRNA-derived stress-induced RNA (tiRNA); tsRNA; ventricular remodeling.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
Novel therapeutic insights into pathological cardiac hypertrophy: tRF-16-R29P4PE regulates PACE4 and metabolic pathways.Biochim Biophys Acta Mol Cell Res. 2025 Mar;1872(3):119920. doi: 10.1016/j.bbamcr.2025.119920. Epub 2025 Feb 11. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 39947523
-
Gene expression profiling of hypertrophic cardiomyocytes identifies new players in pathological remodelling.Cardiovasc Res. 2021 May 25;117(6):1532-1545. doi: 10.1093/cvr/cvaa233. Cardiovasc Res. 2021. PMID: 32717063 Free PMC article.
-
Adiponectin mediates cardioprotection in oxidative stress-induced cardiac myocyte remodeling.Am J Physiol Heart Circ Physiol. 2011 Sep;301(3):H984-93. doi: 10.1152/ajpheart.00428.2011. Epub 2011 Jun 10. Am J Physiol Heart Circ Physiol. 2011. PMID: 21666115 Free PMC article.
-
Melatonin alleviates angiotensin-II-induced cardiac hypertrophy via activating MICU1 pathway.Aging (Albany NY). 2020 Nov 26;13(1):493-515. doi: 10.18632/aging.202159. Epub 2020 Nov 26. Aging (Albany NY). 2020. PMID: 33259334 Free PMC article.
-
Unveiling the role of tRNA-derived small RNAs in MAPK signaling pathway: implications for cancer and beyond.Front Genet. 2024 Mar 26;15:1346852. doi: 10.3389/fgene.2024.1346852. eCollection 2024. Front Genet. 2024. PMID: 38596214 Free PMC article. Review.
References
-
- Nakamura M, Sadoshima J. Mechanisms of physiological and pathological cardiac hypertrophy. Nature reviews Cardiology. 2018;15:387–407. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

