Comparative analysis of duck Tembusu virus Cluster 1 and Cluster 2.1 in Culex tritaeniorhynchus: Insights into viral characteristics, infectivity, and innate immune response
- PMID: 40521371
- PMCID: PMC12166833
- DOI: 10.1016/j.crpvbd.2025.100274
Comparative analysis of duck Tembusu virus Cluster 1 and Cluster 2.1 in Culex tritaeniorhynchus: Insights into viral characteristics, infectivity, and innate immune response
Abstract
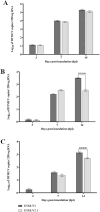
The disease caused by the duck Tembusu virus (DTMUV) is one of the most prevalent arthropod-borne viral diseases in poultry. DTMUV is classified into three distinct clusters based on significant genetic divergence: Cluster 1, Cluster 2 (subdivided into 2.1 and 2.2), and Cluster 3. The virulence of DTMUV in ducks is potentially associated with the virus genotype. The evaluation of different clusters of DTMUV is based predominantly on the characterization of infected duck hosts, and limited attention has been paid to understanding viral virulence toward the infected mosquito vectors. In this study, we explore the infectivity patterns of DTMUV Cluster 1 (DTMUV 1) and Cluster 2.1 (DTMUV 2.1) in the primary mosquito vector, Culex tritaeniorhynchus. Our objective was to explore the relationship between the mosquito vector and DTMUV genotype, intending to determine whether the mosquito vector alters viral biology, thereby influencing the consequential infectivity characteristics in the host cells. We found that variation in viral nonstructural protein-5 (an RNA-dependent RNA polymerase) may influence the antigenicity process in Cx. tritaeniorhynchus. Our results revealed DTMUV1 underwent higher replication than DTMUV2.1 in mosquito salivary glands and saliva. Furthermore, DTMUV1 derived from mosquito saliva produced larger plaque sizes in baby hamster kidney-21 (BHK-21) cells than DTMUV2.1 derived from mosquito saliva. Interestingly, DTMUV2.1 was more efficient than DTMUV1 in inducing the production of mRNAs for macroglobulin complement-related factor, thioester-containing protein, and antimicrobial peptides (cecropin family) within the mosquito salivary gland. Our findings collectively suggest that Cx. tritaeniorhynchus can influence an environment conducive to modifying the amino acid composition of DTMUV1 and DTMUV2.1 in a manner that may affect the innate immune response, consequently augmenting viral virulence.
Keywords: BHK-21 cell; Culex tritaeniorhynchus; Duck Tembusu virus; Innate immune response; NS5.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials

