Uncovering protein conformational dynamics within two-component viral biomolecular condensates
- PMID: 40521608
- PMCID: PMC12168090
- DOI: 10.1002/pro.70181
Uncovering protein conformational dynamics within two-component viral biomolecular condensates
Abstract
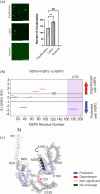
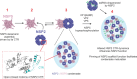
Biomolecular condensates selectively compartmentalize and organize biomolecules within the crowded cellular milieu and are instrumental in some disease mechanisms. Upon infection, many RNA viruses form biomolecular condensates that are often referred to as viral factories. The assembly mechanism of these viral factories remains poorly defined but involves transient, non-stoichiometric protein/RNA interactions, making their structural characterization challenging. Here, we sought to investigate the structural dynamics and intermolecular interactions of the key proteins responsible for condensate formation upon rotavirus infection, namely NSP2 (an RNA chaperone) and NSP5 (an intrinsically disordered protein [IDP]), using a combination of hydrogen-deuterium exchange mass spectrometry (HDX-MS), native MS, and biophysical tools. Our data reveal key structural features of intrinsically disordered NSP5 that are vital for condensate assembly and highlight inter/intra-protein interactions involved in condensate assembly. Moreover, we demonstrate that within a condensate there are altered conformational dynamics within the C-terminal region of NSP2, which has previously been shown to play a role in regulating its RNA chaperoning activity, and in the disordered regions of NSP5. We propose that altered conformational dynamics in NSP2 and NSP5 are critical for regulation of RNA annealing within a biomolecular condensate and for condensate assembly/client recruitment, respectively. Combined, our data demonstrate that the unique environment within a biomolecular condensate can tune functionally important protein conformational dynamics, which may play a crucial role in the replication of rotaviruses.
Keywords: biomolecular condensates; hydrogen–deuterium exchange mass spectrometry; native mass spectrometry; protein dynamics; rotavirus.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

