Novel ATR/PARP1 Dual Inhibitors Demonstrate Synergistic Antitumor Efficacy in Triple-Negative Breast Cancer Models
- PMID: 40521858
- PMCID: PMC12362783
- DOI: 10.1002/advs.202501916
Novel ATR/PARP1 Dual Inhibitors Demonstrate Synergistic Antitumor Efficacy in Triple-Negative Breast Cancer Models
Abstract
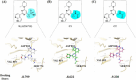
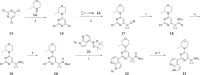
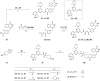
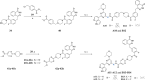
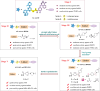
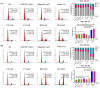
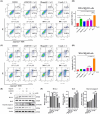
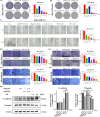
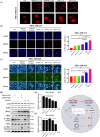
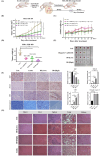
Concomitant inhibition of ataxia telangiectasia and Rad3-related protein (ATR) and poly ADP-ribose Polymerase (PARP) pathways is a promising strategy in cancer therapy, potentially expanding the clinical utility of ATR inhibitor (ATRi) and PARP inhibitor (PARPi). A novel series of ATR/PARP1 dual inhibitors is developed through the pharmacophore fusion of AZD6738 and Olaparib. Among them, B8 emerges as the most promising candidate, exhibiting potent ATR (IC50: 17.3 nM) and PARP1 (IC50: 0.38 nM) inhibition. B8 effectively reduced cell viability, induced apoptosis, and caused G2/M cell cycle arrest in TNBC cells. Additionally, B8 significantly impaired TNBC colony formation, migration, and invasion. Mechanistically, B8 induces DNA damage, evidenced by increased γH2AX levels. In in vivo studies, B8 suppressed tumor growth more effectively than the combination in MDA-MB-468 xenografted mice, with no significant body weight loss. B8 also enhanced γH2AX expression in tumor tissues. These findings confirm the synergistic effects of ATR/PARP1 co-inhibition and highlight the potential of this novel inhibitor class for TNBC therapy.
Keywords: ATR; Anticancer; Dual Inhibitor; PARP1; TNBC.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
All authors declare no competing interests.
Figures








References
-
- Yi Y. W., Int. J. Mol. Sci. 2023, 24, 3704.
MeSH terms
Substances
Grants and funding
- 2023ZY1014/Zhejiang Provincial Department of Science and Technology
- 2023KY210/Medical Health Science and Technology Project of Zhejiang Provincial Health Commission
- G2021017004/Ministry of Science and Technology of China (High-end foreign experts' program
- G20200217005/Ministry of Science and Technology of China (High-end foreign experts' program
- 202412171/Hangzhou's "115" plan to introduce overseas intelligence projects
LinkOut - more resources
Full Text Sources
Miscellaneous
