Amyloid formation of alternatively spliced variants of α-synuclein
- PMID: 40522183
- PMCID: PMC12168488
- DOI: 10.1002/pro.70195
Amyloid formation of alternatively spliced variants of α-synuclein
Abstract
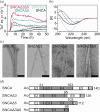
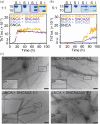
Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy are disorders characterized by the presence of cytosolic α-synuclein (SNCA) amyloids. The gene SNCA is alternatively spliced, generating three variants of SNCA, missing exon 3 (SNCAΔ3) or 5 (SNCAΔ5), or both exons (SNCAΔ3Δ5). Despite purported upregulation in disease states, their pathological relevance is ill-defined. Here, we investigated the amyloid formation of alternatively spliced variants under physiological conditions. Aggregation kinetics, secondary structure, and fibril morphology of N-terminally acetylated SNCAΔ3, SNCAΔ5, and SNCAΔ3Δ5 were assessed by thioflavin-T fluorescence, circular dichroism spectroscopy, and transmission electron microscopy, respectively. Compared to SNCA, both SNCAΔ5 and SNCAΔ3Δ5 aggregate faster and adopt a more twisted fibril morphology, whereas SNCAΔ3 is more sensitive to solution conditions, exhibiting similar or modestly faster aggregation kinetics compared to SNCA. Cross-seeding experiments using spliced-variant fibrils and soluble SNCA showed that despite fibril morphological differences, SNCAΔ5 were competent seeds for SNCA, which is explained by their similar protease-K resistant regions. Contrastingly, neither SNCAΔ3 nor SNCAΔ3Δ5 fibrils cross-seed SNCA, indicating exon 3 (residues 41-54) is essential in modulating fibril structure. Notably, SNCA aggregation is stimulated by sub-stoichiometric amounts of soluble SNCAΔ5 and SNCAΔ3Δ5, but not SNCAΔ3, suggesting that exon 5 (residues 103-130) is more important in modulating aggregation kinetics. Taken together, we propose that alternatively spliced variants are pathogenic by exacerbating aggregation of the main SNCA isoform.
Keywords: Parkinson's disease; aggregation; alternative splicing; amyloid; circular dichroism; electron microscopy; fibril; kinetics; α‐synuclein.
Published 2025. This article is a U.S. Government work and is in the public domain in the USA. Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, et al. Phosphorylation of Ser‐129 is the dominant pathological modification of α‐synuclein in familial and sporadic Lewy body disease. J Biol Chem. 2006;281:29739–29752. - PubMed
-
- Apffel A, Fischer S, Goldberg G, Goodley PC, Kuhlmann FE. Enhanced sensitivity for peptide mapping with electrospray liquid chromatography‐mass spectrometry in the presence of signal suppression due to trifluoroacetic acid‐containing mobile phases. J Chromatogr A. 1995;712:177–190. - PubMed
-
- Bell R, Castellana‐Cruz M, Nene A, Thrush RJ, Xu CK, Kumita JR, et al. Effects of N‐terminal acetylation on the aggregation of disease‐related α‐synuclein variants. J Mol Biol. 2023;435:167825. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

