Core Flipping in Lead Optimization: Rank Ordering Using λ-Dynamics
- PMID: 40522341
- PMCID: PMC12264938
- DOI: 10.1021/acs.jcim.5c00320
Core Flipping in Lead Optimization: Rank Ordering Using λ-Dynamics
Abstract
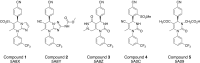
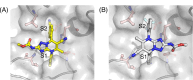
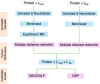
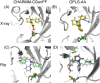
In structure-based drug discovery, reliable structural models of ligands bound to their target receptors are critical for establishing the structure-activity relationship of the congeneric series. In such a series, substitutions on a common scaffold core might lead to different binding modes, ranging from slight changes of orientations to flipping or inversion of the core structure. Moreover, molecular docking might lead to alternative orientations within the top-ranked poses without being able to discriminate which is most likely. To determine the relative binding affinities between two alternative ligand poses, we propose a methodology based on relative binding free energy calculations using the λ-dynamics method. We used a dual-topology approach with distance-restraining schemes. We introduced a novel strategy using a one-step perturbation to calculate the contributions of the applied restraints. While using FEP/MBAR instead for that purpose led to smaller uncertainties, it suffered from convergence issues. We tested the validity and predictive power of our approach using two pharmaceutically relevant targets and eight small-molecule inhibitors from the experimentally characterized congeneric series. For each target, our approach correctly ranks the known X-ray poses as more favorable than alternative flipped poses. The proposed methodology can be easily extended to rank more than two poses and should also be applicable to the evaluation of alternative rotamers of target amino acids.
Figures

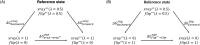
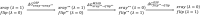
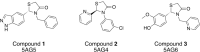

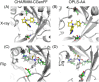

References
-
- Wang L., Wu Y., Deng Y., Kim B., Pierce L., Krilov G., Lupyan D., Robinson S., Dahlgren M. K., Greenwood J.. et al. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J. Am. Chem. Soc. 2015;137:2695–2703. doi: 10.1021/ja512751q. - DOI - PubMed
-
- Schindler C. E. M., Baumann H., Blum A., Böse D., Buchstaller H.-P., Burgdorf L., Cappel D., Chekler E., Czodrowski P., Dorsch D.. et al. Large-Scale Assessment of Binding Free Energy Calculations in Active Drug Discovery Projects. J. Chem. Inf. Model. 2020;60:5457–5474. doi: 10.1021/acs.jcim.0c00900. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

