Transcriptomic profiling of pancreatic neuroendocrine tumors: dysregulation of WNT, MAPK, PI3K, neddylation pathways and potential non-invasive biomarkers
- PMID: 40522948
- PMCID: PMC12169574
- DOI: 10.1371/journal.pone.0325672
Transcriptomic profiling of pancreatic neuroendocrine tumors: dysregulation of WNT, MAPK, PI3K, neddylation pathways and potential non-invasive biomarkers
Abstract
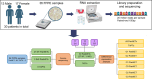
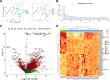
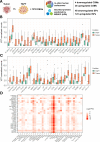
The study aimed to identify altered signaling pathways and potential non-invasive biomarkers for pancreatic neuroendocrine tumors (PanNETs) through transcriptomic profiling of tumor tissues. The analysis encompassed samples from non-functional PanNETs (NF-PanNETs), insulinomas, and tumor-adjacent pancreatic tissues (TAPT). In the differential expression analysis comparing PanNETs and TAPTs, we identified 1,210 differentially expressed genes at a false discovery rate significance threshold of < 0.05 and with Log2FoldChange values of > 0.5 and <-0.5. Further pathway enrichment analysis revealed a multitude of overrepresented signaling pathways related to cell proliferation, survival, and tumorigenesis. Significant findings included the Beta-catenin-independent and TCF-dependent WNT signaling pathways, MAPK1/MAPK3 signaling, and terms associated with PI3K/AKT/mTOR signaling. Among the list of DEGs, we also identified 28 upregulated genes encoding cell surface proteins and 24 upregulated genes encoding cancer-associated secretome proteins. Since the proteins of these genes are found in the bloodstream, there is potential for further testing of these markers as biomarkers for liquid biopsy assays. Overall, these findings underscore the promise of transcriptomic landscape analysis in identifying PanNET-specific non-invasive biomarkers and uncovering potential therapeutic targets.
Copyright: © 2025 Niedra et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
miRNA profile in pancreatic neuroendocrine tumors: Preliminary results.Sci Prog. 2025 Jan-Mar;108(1):368504251326864. doi: 10.1177/00368504251326864. Epub 2025 Mar 28. Sci Prog. 2025. PMID: 40152231 Free PMC article.
-
CEP55, A Promising Prognostic Biomarker for Pancreatic Neuroendocrine Neoplasms, Promotes Tumor Progression Through Activation of PI3K/AKT/mTOR Pathway.FASEB J. 2025 Apr 30;39(8):e70535. doi: 10.1096/fj.202402990R. FASEB J. 2025. PMID: 40249668
-
Deep sequencing reveals distinct microRNA-mRNA signatures that differentiate pancreatic neuroendocrine tumor from non-diseased pancreas tissue.BMC Cancer. 2025 Apr 11;25(1):669. doi: 10.1186/s12885-025-14043-w. BMC Cancer. 2025. PMID: 40217502 Free PMC article.
-
Diagnostic and prognostic biomarkers for pancreatic neuroendocrine neoplasms.Pathologie (Heidelb). 2024 Nov;45(Suppl 1):74-82. doi: 10.1007/s00292-024-01393-8. Epub 2024 Nov 18. Pathologie (Heidelb). 2024. PMID: 39556246 Review. English.
-
The landscape of molecular alterations in pancreatic and small intestinal neuroendocrine tumours.Ann Endocrinol (Paris). 2019 Jun;80(3):153-158. doi: 10.1016/j.ando.2019.04.010. Epub 2019 Apr 11. Ann Endocrinol (Paris). 2019. PMID: 31072588 Review.
References
-
- Young K, Lawlor RT, Ragulan C, Patil Y, Mafficini A, Bersani S, et al. Immune landscape, evolution, hypoxia-mediated viral mimicry pathways and therapeutic potential in molecular subtypes of pancreatic neuroendocrine tumours. Gut. 2021;70(10):1904–13. doi: 10.1136/gutjnl-2020-321016 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

