Influence of multi-species data on gene-disease associations in substance use disorder using random walk with restart models
- PMID: 40522980
- PMCID: PMC12169588
- DOI: 10.1371/journal.pone.0325201
Influence of multi-species data on gene-disease associations in substance use disorder using random walk with restart models
Abstract
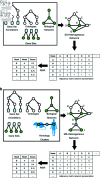
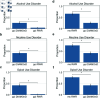
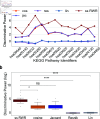
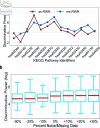
A major challenge lies in discovering, emphasizing, and characterizing human gene-disease and gene-gene associations. The limitations of data on the role of human gene products in substance use disorder (SUD) makes it challenging to transition from genetic associations to actionable insights. The integration of data from multiple diverse sources, including information-dense studies in model organisms, has the potential to address this gap. We demonstrate a modified performance of the Random Walk with Restart algorithm when multi-species data is integrated in the heterogeneous network within the context of SUD. Additionally, our approach distinguishes among disparate pathways derived from the Kyoto Encyclopedia of Genes and Genomes. Thus, we conclude that direct incorporation of multi-species data to an aggregated heterogeneous knowledge graph can adjust RWR's performance and enables users to discover new gene-disease and gene-gene associations.
Copyright: © 2025 Castaneda et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hatoum AS, Colbert SMC, Johnson EC, Huggett SB, Deak JD, Pathak G, et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Nat Ment Health. 2023;1(3):210–23. doi: 10.1038/s44220-023-00034-y - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

