A distinct LHCI arrangement is recruited to photosystem I in Fe-starved green algae
- PMID: 40523173
- PMCID: PMC12207447
- DOI: 10.1073/pnas.2500621122
A distinct LHCI arrangement is recruited to photosystem I in Fe-starved green algae
Abstract
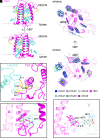
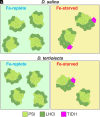
Iron (Fe) availability limits photosynthesis at a global scale where Fe-rich photosystem (PS) I abundance is drastically reduced in Fe-poor environments. We used single-particle cryoelectron microscopy to reveal a unique Fe starvation-dependent arrangement of light-harvesting chlorophyll (LHC) proteins where Fe starvation-induced TIDI1 is found in an additional tetramer of LHC proteins associated with PSI in Dunaliella tertiolecta and Dunaliella salina. These cosmopolitan green algae are resilient to poor Fe nutrition. TIDI1 is a distinct LHC protein that co-occurs in diverse algae with flavodoxin (an Fe-independent replacement for the Fe-containing ferredoxin). The antenna expansion in eukaryotic algae we describe here is reminiscent of the iron-starvation induced (isiA-encoding) antenna ring in cyanobacteria, which typically co-occurs with isiB, encoding flavodoxin. Our work showcases the convergent strategies that evolved after the Great Oxidation Event to maintain PSI capacity.
Keywords: TIDI; iron homeostasis; iron starvation–induced protein A (isiA); phytoplankton; structural biology.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P., Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281, 237–240 (1998). - PubMed
-
- Chen Y., Barak P., “Iron nutrition of plants in calcareous soils” in Advances in Agronomy (Elsevier, 1982), pp. 217–240.
-
- Moore C. M., et al. , Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710 (2013).
-
- Raven J. A., Predictions of Mn and Fe use efficiencies of phototrophic growth as a function of light availability for growth and of C assimilation pathway. New Phytol. 116, 1–18 (1990).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

