hnRNP A1 induces aberrant CFTR exon 9 splicing via a newly discovered ESS element
- PMID: 40523798
- PMCID: PMC12171016
- DOI: 10.26508/lsa.202402720
hnRNP A1 induces aberrant CFTR exon 9 splicing via a newly discovered ESS element
Abstract
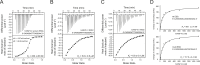
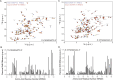
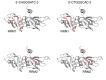
RNA-protein interactions play a key role in the aberrant splicing of CFTR exon 9. Exon 9 skipping leads to the production of a nonfunctional chloride channel associated with severe forms of cystic fibrosis. The missplicing depends on TDP-43 binding to an extended UG-rich binding site upstream of CFTR exon 9 3' splicing site (3'ss) and is associated with concomitant hnRNP A1 recruitment. Although TDP-43 is the dominant inhibitor of exon 9 inclusion, the role of hnRNP A1, a protein with two RNA recognition motifs, remained unclear. In this work, we have studied the interaction between hnRNP A1 and the CFTR pre-mRNA using NMR spectroscopy and Isothermal Titration Calorimetry. The affinities are submicromolar, and Isothermal Titration Calorimetry data suggest complexes with a 1:1 stoichiometry. NMR titrations reveal that hnRNP A1 interacts with model CTFR 3'ss sequences in a fast exchange regime at the NMR timescale. Splicing assays finally show that this hnRNP A1 binding site represents a previously unknown exonic splicing silencer element. Together, our results shed light on the mechanism of aberrant CFTR exon 9 splicing.
© 2025 Beaumont et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
