No evidence that human GIGYF2 interacts with GRB10: implications for human disease
- PMID: 40523800
- PMCID: PMC12171015
- DOI: 10.26508/lsa.202503334
No evidence that human GIGYF2 interacts with GRB10: implications for human disease
Abstract
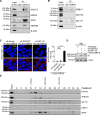
GIGYF2 (growth factor receptor-bound protein 10 [GRB10]-interacting GYF [glycine-tyrosine-phenylalanine] protein 2) reduces mRNA stability and translation via microRNAs, ribosome quality control, and several RNA-binding proteins. GIGYF2 was first identified in mouse cell lines as an interacting partner with GRB10, which binds to the insulin receptor and the insulin-like growth factor receptor 1. Mutations in the human GIGYF2 gene were reported in autism. In mouse models, Gigyf2 mutations engender several diseases. It was therefore thought that the GIGYF2-associated disease in humans is caused by defective GRB10 signaling. We show here that GIGYF2 does not interact with GRB10 in human cell lines, as determined by co-immunoprecipitation and proximity ligation assays. The lack of interaction is explained by the absence of the critical GYF domain-binding PPGΦ sequence in the human GRB10 protein. These results contrast with the current understanding that a GIGYF2/GRB10 complex is associated with human disease via insulin receptor and insulin-like growth factor receptor 1 signaling and underscore alternative mechanisms responsible for the observed phenotypes associated with mutations in the human GIGYF2 gene.
© 2025 Choi et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
