Repurposing MDM2 inhibitor RG7388 for TP53-mutant NSCLC: a p53-independent pyroptotic mechanism via ROS/p-p38/NOXA/caspase-3/GSDME axis
- PMID: 40523886
- PMCID: PMC12170848
- DOI: 10.1038/s41419-025-07770-2
Repurposing MDM2 inhibitor RG7388 for TP53-mutant NSCLC: a p53-independent pyroptotic mechanism via ROS/p-p38/NOXA/caspase-3/GSDME axis
Abstract
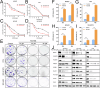
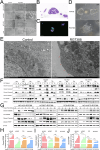
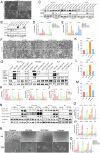
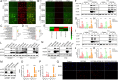
Non-small cell lung cancer (NSCLC) is highly malignant with limited treatment options, largely due to the inherent tumoral heterogeneity and acquired resistance towards chemotherapy and immunotherapy. RG7388, a known MDM2 inhibitor, exhibited anticancer activity in TP53-wild-type (TP53WT) NSCLC by triggering the p53/PUMA axis-dependent apoptosis. However, our study uncovered previously unrecognized p53-independent anticancer effects of RG7388 in TP53-mutant (TP53mutant) NSCLC, although the underlying mechanisms remained elusive. Here, we demonstrated that RG7388 specifically induced the NOXA/caspase-3 axis-dependent apoptosis and gasdermin E (GSDME)-mediated secondary pyroptosis in TP53mutant NSCLC, as validated through in silico analyses and multiple biological assays. Mechanically, we identified reactive oxygen species (ROS) as the critical mediator in NOXA upregulation and p38 MAPK pathway activation in RG7388 treated TP53mutant NSCLC. This was further supported by the use of ROS scavengers, N-acetylcysteine (NAC), and Ferrostatin-1 (Fer-1), which attenuated these effects. Pharmacologic inhibition of p38 MAPK signaling by SB203580 rescued RG7388-induced ROS-dependent NOXA accumulation and subsequent apoptosis and pyroptosis, highlighting the central role of the ROS/phosphorylated p38 MAPK (p-p38)/NOXA/caspase-3 axis in RG7388-induced TP53mutant NSCLC cell death. Our findings revealed a novel mechanism for selectively targeting mutant p53-derived cancer through ROS/p-p38-mediated NOXA accumulation, offering potential therapeutic implications given the current lack of direct mutant p53 targeting strategies in cancer. Furthermore, immunohistochemical (IHC) analysis of an NSCLC tissue microarray confirmed a strong positive correlation between p-p38 and NOXA expression. Clinical data analysis further suggested that the p-p38/NOXA axis might be a potential prognostic biomarker for overall survival (OS) in NSCLC patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Meyer ML, Fitzgerald BG, Paz-Ares L, Cappuzzo F, Jänne PA, Peters S, et al. New promises and challenges in the treatment of advanced non-small-cell lung cancer. Lancet. 2024;404:803–22. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

