Integration of MRI radiomics and germline genetics to predict the IDH mutation status of gliomas
- PMID: 40523904
- PMCID: PMC12170908
- DOI: 10.1038/s41698-025-00980-z
Integration of MRI radiomics and germline genetics to predict the IDH mutation status of gliomas
Abstract
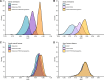
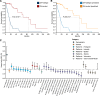
The molecular profiling of gliomas for isocitrate dehydrogenase (IDH) mutations currently relies on resected tumor samples, highlighting the need for non-invasive, preoperative biomarkers. We investigated the integration of glioma polygenic risk scores (PRS) and radiographic features for prediction of IDH mutation status. We used 256 radiomic features, a glioma PRS and demographic information in 158 glioma cases within elastic net and neural network models. The integration of glioma PRS with radiomics increased the area under the receiver operating characteristic curve (AUC) for distinguishing IDH-wildtype vs. IDH-mutant glioma from 0.83 to 0.88 (PΔAUC = 6.9 × 10-5) in the elastic net model and from 0.91 to 0.92 (PΔAUC = 0.32) in the neural network model. Incorporating age at diagnosis and sex further improved the classifiers (elastic net: AUC = 0.93, neural network: AUC = 0.93). Patients predicted to have IDH-mutant vs. IDH-wildtype tumors had significantly lower mortality risk (hazard ratio (HR) = 0.18, 95% CI: 0.08-0.40, P = 2.1 × 10-5), comparable to prognostic trajectories for biopsy-confirmed IDH status. The augmentation of imaging-based classifiers with genetic risk profiles may help delineate molecular subtypes and improve the timely, non-invasive clinical assessment of glioma patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures