Recent advances in deep learning for protein-protein interaction: a review
- PMID: 40524189
- PMCID: PMC12168265
- DOI: 10.1186/s13040-025-00457-6
Recent advances in deep learning for protein-protein interaction: a review
Abstract
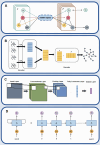
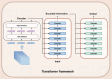
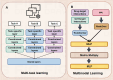
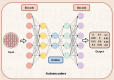
Deep learning, a cornerstone of artificial intelligence, is driving rapid advancements in computational biology. Protein-protein interactions (PPIs) are fundamental regulators of biological functions. With the inclusion of deep learning in PPI research, the field is undergoing transformative changes. Therefore, there is an urgent need for a comprehensive review and assessment of recent developments to improve analytical methods and open up a wider range of biomedical applications. This review meticulously assesses deep learning progress in PPI prediction from 2021 to 2025. We evaluate core architectures (GNNs, CNNs, RNNs) and pioneering approaches-attention-driven Transformers, multi-task frameworks, multimodal integration of sequence and structural data, transfer learning via BERT and ESM, and autoencoders for interaction characterization. Moreover, we examined enhanced algorithms for dealing with data imbalances, variations, and high-dimensional feature sparsity, as well as industry challenges (including shifting protein interactions, interactions with non-model organisms, and rare or unannotated protein interactions), and offered perspectives on the future of the field. In summary, this review systematically summarizes the latest advances and existing challenges in deep learning in the field of protein interaction analysis, providing a valuable reference for researchers in the fields of computational biology and deep learning.
Keywords: Artificial intelligence; Artificial neural networks; Computational biology; Deep learning; Machine learning; PPI prediction; Protein-protein interactions.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
The ultimate power play in research - partnering with patients, partnering with power.Res Involv Engagem. 2025 Jun 17;11(1):65. doi: 10.1186/s40900-025-00745-9. Res Involv Engagem. 2025. PMID: 40528262 Free PMC article.
-
Prediction of Anti-rheumatoid Arthritis Natural Products of Xanthocerais Lignum Based on LC-MS and Artificial Intelligence.Comb Chem High Throughput Screen. 2025;28(4):627-646. doi: 10.2174/0113862073282138240116112348. Comb Chem High Throughput Screen. 2025. PMID: 38299408 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Emerging research trends in artificial intelligence for cancer diagnostic systems: A comprehensive review.Heliyon. 2024 Aug 23;10(17):e36743. doi: 10.1016/j.heliyon.2024.e36743. eCollection 2024 Sep 15. Heliyon. 2024. Retraction in: Heliyon. 2025 Apr 15;11(9):e43335. doi: 10.1016/j.heliyon.2025.e43335. PMID: 39263113 Free PMC article. Retracted. Review.
-
Artificial intelligence methods applied to longitudinal data from electronic health records for prediction of cancer: a scoping review.BMC Med Res Methodol. 2025 Jan 28;25(1):24. doi: 10.1186/s12874-025-02473-w. BMC Med Res Methodol. 2025. PMID: 39875808 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

