Integrative analysis of bulk and single-cell gene expression profiles to identify bone marrow mesenchymal cell heterogeneity and prognostic significance in multiple myeloma
- PMID: 40524208
- PMCID: PMC12172380
- DOI: 10.1186/s12967-025-06637-6
Integrative analysis of bulk and single-cell gene expression profiles to identify bone marrow mesenchymal cell heterogeneity and prognostic significance in multiple myeloma
Abstract
Background: Multiple myeloma is a hematologic malignancy characterized by complex interactions within the tumor microenvironment, where mesenchymal stem cells (MSCs) contribute significantly to disease progression, immune suppression, and drug resistance.
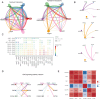
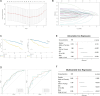
Methods: This study investigated the heterogeneity of MSCs in multiple myeloma using single-cell RNA sequencing (10X) and bulk transcriptomic data. Further analysis was performed by Seurat, SCENIC, CellChat. GSE4581 and GSE136337 were used as training set and validation set to construct a newly described prognostic model through COX and LASSO.
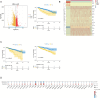
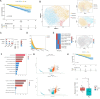
Results: By analyzing bone marrow samples from healthy donors and multiple myeloma patients at different Revised International Staging System (R-ISS) stages, this study identified distinct MSC subpopulations, including osteogenic, angiogenic, immune regulatory, and multipotent clusters, each of which plays unique roles in the tumor microenvironment. Interestingly, we found a unique subclone with upregulated expression of high mobility group proteins, these MSC exert a strong regulatory effect, which was defined as "HMGhMSC".
Conclusions: Our findings reveal that the proportion of osteogenic MSCs, which are crucial for bone health, decreases as the disease progresses, which is correlated with the bone lysis commonly observed in advanced multiple myeloma. Additionally, immune regulatory MSCs contribute to the formation of an immunosuppressive microenvironment, promoting tumor immune evasion. A prognostic model based on HMGhMSC subpopulations was developed, which demonstrated that these cells have significant potential as therapeutic targets for improving the prognosis and developing treatments for bone disease in multiple myeloma patients.
Keywords: High mobility group proteins; Immunosuppressive microenvironment; Mesenchymal stem cells; Multiple myeloma; Osteogenesis; Prognostic model; Single-cell RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: No potential conflicts of interest were disclosed by all authors.
Figures


References
-
- Dufva O, et al. Single-cell functional genomics reveals determinants of sensitivity and resistance to natural killer cells in blood cancers. Immunity. 2023;56:2816–35. 10.1016/j.immuni.2023.11.008. - PubMed
-
- Ledergor G, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med. 2018;24:1867–76. 10.1038/s41591-018-0269-2. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

