Hub Genes PRPF19 and PPIB: Molecular Pathways and Potential Biomarkers in COPD
- PMID: 40524719
- PMCID: PMC12168939
- DOI: 10.2147/COPD.S511696
Hub Genes PRPF19 and PPIB: Molecular Pathways and Potential Biomarkers in COPD
Abstract
Background: Chronic Obstructive Pulmonary Disease (COPD), a complex respiratory disorder, results from genetic and environmental factors. Uncovering its genetic basis is vital for diagnostics and treatment. Robust genetic analysis is essential to establish a causal link.
Methods: Genome-wide DNA methylation analysis was performed using the Illumina Infinium HumanMethylation850 BeadChip in peripheral blood from 8 COPD patients and 8 healthy smoking controls. Differentially methylated genes (DMGs) were cross-analyzed with differentially expressed genes (DEGs) identified from the Gene Expression Omnibus (GEO) dataset GSE38974 (23 COPD, 9 controls). Weighted gene co-expression network analysis (WGCNA) and protein-protein interaction (PPI) networks were utilized to identify COPD-associated hub genes. Mendelian randomization (MR) analysis examined the causal relationship between hub genes and COPD. The expression of selected hub genes was validated through RT-qPCR (80 COPD, 62 controls), immunohistochemistry, and Western blot analyses (10 COPD and 10 controls).
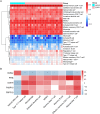
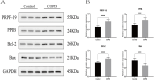
Results: We found 10,593 DMGs and 646 DEGs associated with COPD. These genes were compared with WGCNA module genes, and the Protein-Protein Interaction (PPI) network interaction diagram was drawn, thereby identifying five Hub genes: PPIB, HSPA2, PRPF19, FKBP10 and DOHH. The expression levels of DOHH, FKBP10, PPIB and PRPF19 are higher in COPD, while the expression level of HSPA2 is lower. MR results indicate a potential causal relationship between PRPF19, PPIB and COPD. RT-qPCR, immunohistochemistry and Western blot experiments verified that the expression of PRPF-19 and PPIB was up-regulated in peripheral blood and lung tissue, which was consistent with the results of bioinformatics analysis.
Conclusion: Our findings suggest that PRPF19 and PPIB may serve as promising diagnostic biomarkers in COPD. Further studies are required to fully elucidate their roles in COPD pathogenesis.
Keywords: Mendelian randomization; chronic obstructive pulmonary disease; epigenetic susceptibility; hub genes; protein-protein interaction.
© 2025 Zhao et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures





Similar articles
-
Screening and Verification COPD-OSA Overlap Syndrome Core Genes Using Bioinformatics.Int J Chron Obstruct Pulmon Dis. 2025 Jul 26;20:2601-2614. doi: 10.2147/COPD.S528703. eCollection 2025. Int J Chron Obstruct Pulmon Dis. 2025. PMID: 40747360 Free PMC article.
-
Developing a Panel of Shared Susceptibility Genes as Diagnostic Biomarkers for chronic obstructive pulmonary disease and Heart Failure.Comput Biol Med. 2025 Sep;196(Pt A):110657. doi: 10.1016/j.compbiomed.2025.110657. Epub 2025 Jul 4. Comput Biol Med. 2025. PMID: 40617086
-
Identification of hub genes and prediction of the ceRNA network in adult sepsis.PeerJ. 2025 Aug 13;13:e19619. doi: 10.7717/peerj.19619. eCollection 2025. PeerJ. 2025. PMID: 40821971 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
A bibliometric analysis of research trends in mesenchymal stem cell therapy for neonatal bronchopulmonary dysplasia: 2004-2024.Front Pediatr. 2025 Jun 3;13:1558301. doi: 10.3389/fped.2025.1558301. eCollection 2025. Front Pediatr. 2025. PMID: 40530182 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

