Prevalence and environmental abundance of the TSET complex in cosmopolitan algal groups
- PMID: 40524958
- PMCID: PMC12167817
- DOI: 10.1016/j.isci.2025.112679
Prevalence and environmental abundance of the TSET complex in cosmopolitan algal groups
Abstract
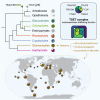
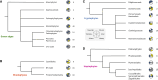
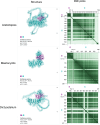
Classical cell biology paradigms are largely established on animal, fungal, and plant models which constitute a small fraction of eukaryotic diversity. Some important cellular machinery has been historically overlooked due to their absence from animals and fungi, e.g., the membrane-trafficking complex TSET involved in plant cell division and endocytosis. Here, we document TSET complexes in distantly related photosynthetic eukaryotic groups (green algae, red algae, haptophytes, cryptophytes, and stramenopiles including diatoms). 3D modeling predicts that at least some stramenopile-encoded subunits share conserved structural features with plant orthologues, and gene expression analysis from the diatom Phaeodactylum tricornutum shows that they are co-expressed with endomembrane trafficking proteins. Finally, diatom TSET genes are detectable in meta-transcriptomic data from Tara Oceans, suggesting functional roles in the wild. These results support the importance of integrating non-model organisms into our understanding of eukaryotic cell biology, as they may reveal underappreciated protein complexes essential for cellular and ecosystem functions.
Keywords: Aquatic biology; Cell biology; Plant biology; Plant development; Plant evolution.
© 2025 The Authors.
Conflict of interest statement
The authors have no competing interests to declare.
Figures




Similar articles
-
Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution.PLoS One. 2011;6(12):e29138. doi: 10.1371/journal.pone.0029138. Epub 2011 Dec 14. PLoS One. 2011. PMID: 22195008 Free PMC article.
-
Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae.BMC Evol Biol. 2015 Feb 10;15:16. doi: 10.1186/s12862-015-0286-4. BMC Evol Biol. 2015. PMID: 25887237 Free PMC article.
-
Characterization of a Marine Diatom Chitin Synthase Using a Combination of Meta-Omics, Genomics, and Heterologous Expression Approaches.mSystems. 2023 Apr 27;8(2):e0113122. doi: 10.1128/msystems.01131-22. Epub 2023 Feb 15. mSystems. 2023. PMID: 36790195 Free PMC article.
-
Phaeodactylum tricornutum: An established model species for diatom molecular research and an emerging chassis for algal synthetic biology.J Phycol. 2023 Dec;59(6):1114-1122. doi: 10.1111/jpy.13400. Epub 2023 Nov 17. J Phycol. 2023. PMID: 37975560 Review.
-
Molecular mechanisms of endomembrane trafficking in plants.Plant Cell. 2022 Jan 20;34(1):146-173. doi: 10.1093/plcell/koab235. Plant Cell. 2022. PMID: 34550393 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

