Single-Cell RNA Sequencing Delineates Renal Anti-Fibrotic Mechanisms Mediated by TRPC6 Inhibition
- PMID: 40525246
- PMCID: PMC12412467
- DOI: 10.1002/advs.202501175
Single-Cell RNA Sequencing Delineates Renal Anti-Fibrotic Mechanisms Mediated by TRPC6 Inhibition
Abstract
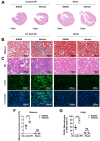
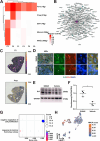
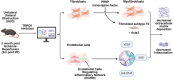
Chronic kidney disease (CKD) is characterized by persistent inflammation and tubulointerstitial fibrosis leading to end-stage renal disease. Transient receptor potential canonical 6 (TRPC6) channel inhibition mitigates tubular injury and renal fibrosis in murine models of unilateral ureteral obstruction (UUO) and 2-month chronic post-ischemia-reperfusion injury (2m post-I/R). Through integrated analysis of single-cell-RNA-sequencing (scRNA-Seq) data from UUO mice treated with the selective TRPC6 inhibitor SH045, here the renoprotective cell composition and cell type-specific transcriptional programs are defined. We explored translational aspects by conducting an in-depth scRNA-Seq analysis of kidney samples from patients with CKD. These results reveal global transcriptional shifts with a dramatic diversification of inflammatory cells, endothelial cells and fibroblasts. Notably, a distinct subpopulation of novel endothelial cells is delineated, which is termed ECRIN, that regulate inflammatory networks implicating VEGF and GAS signaling pathways. The data also indicates that inhibition of TRPC6 channels triggers a Prnp transcription factor regulatory network, which contributes to the alleviation of renal fibrosis. The key findings are supported at the protein level by immunofluorescence and western blot analysis. We observed similar patterns in the chronic 2m postI/R injury model. These findings provide novel insights into the potential therapeutic benefits of TRPC6 inhibition in CKD.
Keywords: chronic kidney disease; renal fibrosis; single‐cell rna sequencing; spatial transcriptomics.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Kalantar‐Zadeh K., Jafar T. H., Nitsch D., Neuen B. L., Perkovic V., Lancet 2021, 398, 786. - PubMed
MeSH terms
Substances
Grants and funding
- GO766/18-2/Deutsche Forschungsgemeinschaft
- GO766/22-2/Deutsche Forschungsgemeinschaft
- GO766/25-1/Deutsche Forschungsgemeinschaft
- GO766/26-1/Deutsche Forschungsgemeinschaft
- SFB1365/Deutsche Forschungsgemeinschaft
- 01GY2201/German Ministry of Education and Research
- FOVB-2023-12/Deutscher Akademischer Austauschdienst (DAAD) and University Medicine Greifswald, Anschubfinanzierung
- NSFC/National Natural Science Foundation of China
- 82070718/National Natural Science Foundation of China
- 23ZR1451000/Shanghai Science and Technology Innovation Natural Foundation
- 24PJD088/Shanghai Pujiang Program
LinkOut - more resources
Full Text Sources
Medical
