Identifying Retinal Features Using a Self-Configuring CNN for Clinical Intervention
- PMID: 40525921
- PMCID: PMC12178434
- DOI: 10.1167/iovs.66.6.55
Identifying Retinal Features Using a Self-Configuring CNN for Clinical Intervention
Abstract
Purpose: Retinal diseases are leading causes of blindness worldwide, necessitating accurate diagnosis and timely treatment. Optical coherence tomography (OCT) has become a universal imaging modality of the retina in the past 2 decades, aiding in the diagnosis of various retinal conditions. However, the scarcity of comprehensive, annotated OCT datasets, that are labor-intensive to assemble, has hindered the advancement of artificial intelligence (AI)-based diagnostic tools.
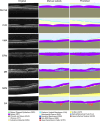
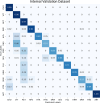
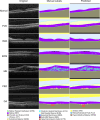
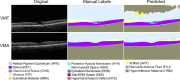
Methods: To address the lack of annotated OCT segmentation datasets, we introduce OCTAVE, an extensive 3D OCT dataset with high-quality, pixel-level annotations for anatomic and pathological structures. Additionally, we provide similar annotations for four independent public 3D OCT datasets, enabling their use as external validation sets. To demonstrate the potential of this resource, we train a deep learning segmentation model using the self-configuring no-new-U-Net (nnU-Net) framework and evaluate its performance across all four external validation sets.
Results: The OCTAVE dataset collected consists of 198 OCT volumes (3762 B-scans) used for training and 221 OCT volumes (4109 B-scans) for external validation. The trained deep learning model demonstrates clinically significant performance across all retinal structures and pathological features.
Conclusions: We demonstrate robust segmentation performance and generalizability across independently collected datasets. OCTAVE bridges the gap in publicly available datasets, supporting the development of AI tools for precise disease detection, monitoring, and treatment guidance. This resource has the potential to improve clinical outcomes and advance AI-driven retinal disease management.
Conflict of interest statement
Disclosure:
Figures




Similar articles
-
A novel deep learning framework for retinal disease detection leveraging contextual and local features cues from retinal images.Med Biol Eng Comput. 2025 Jul;63(7):2029-2046. doi: 10.1007/s11517-025-03314-0. Epub 2025 Feb 7. Med Biol Eng Comput. 2025. PMID: 39918766
-
Joint segmentation of retinal layers and fluid lesions in optical coherence tomography with cross-dataset learning.Artif Intell Med. 2025 Apr;162:103096. doi: 10.1016/j.artmed.2025.103096. Epub 2025 Feb 21. Artif Intell Med. 2025. PMID: 39999658
-
A comparative study of machine learning models for automated detection and classification of retinal diseases in Ghana.PLoS One. 2025 Aug 1;20(8):e0327743. doi: 10.1371/journal.pone.0327743. eCollection 2025. PLoS One. 2025. PMID: 40748964 Free PMC article.
-
The march to harmonized imaging standards for retinal imaging.Prog Retin Eye Res. 2025 Jul;107:101363. doi: 10.1016/j.preteyeres.2025.101363. Epub 2025 May 11. Prog Retin Eye Res. 2025. PMID: 40360070 Review.
-
Is Artificial Intelligence an accurate tool for improving access to ophthalmological services in rural areas? A narrative review.Ann Agric Environ Med. 2025 Jun 27;32(2):320-322. doi: 10.26444/aaem/195109. Epub 2024 Nov 27. Ann Agric Environ Med. 2025. PMID: 40586515 Review.
References
-
- Fung AT, Galvin J, Tran T. Epiretinal membrane: a review. Clin Exp Ophthalmol. 2021; 49(3): 289–308. - PubMed
-
- Wong WL, Su X, Li X, et al.. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014; 2(2): e106–e116. - PubMed
-
- Liu L, Swanson M. Improving patient outcomes: role of the primary care optometrist in the early diagnosis and management of age-related macular degeneration. Clin Optom. 2013; 5: 1–12.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

