Enhancing Clinical Detection Accuracy of Large Structured Viral RNA via DNAzyme Cleavage and Antisense-Assisted Rolling Circle Amplification
- PMID: 40526841
- PMCID: PMC12338419
- DOI: 10.1002/anie.202507973
Enhancing Clinical Detection Accuracy of Large Structured Viral RNA via DNAzyme Cleavage and Antisense-Assisted Rolling Circle Amplification
Abstract
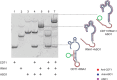
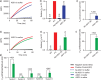
Sensitive detection of viral RNA is critical for accurate diagnostic testing, particularly during outbreaks of emerging infectious diseases. Rolling circle amplification (RCA) is a powerful isothermal amplification strategy that can be directly primed by RNA, eliminating the need for reverse transcription. Previous approaches have used 10-23 DNAzymes to cleave viral RNA, generating 3'-ends for hybridization to circular DNA templates (CDTs). However, the resulting RNA fragments often retained secondary or tertiary structures that hindered CDT binding and limited RCA efficiency. To address this challenge, we developed antisense oligonucleotide-assisted RCA (ASO-RCA), a general strategy that uses short upstream antisense oligonucleotides (ASOs) to remodel RNA structure and expose the CDT-binding site. Using five DNAzyme-CDT systems targeting distinct regions of the SARS-CoV-2 genome, we show that ASO inclusion improves CDT hybridization and enhances RCA output-by up to 70-fold. This enhancement was observed using both linear and quasi-exponential RCA formats and remained effective in 50% pooled saliva. When applied to clinical saliva samples, ASO-assisted RCA markedly improved diagnostic performance, achieving 100% sensitivity and up to 97.5%-100% accuracy across multiple systems. These findings establish ASO-DNAzyme-RCA as a simple, robust, and clinically relevant platform for improving nucleic acid detection in structured RNA targets.
Keywords: Antisense oligonucleotides; Biosensors; DNAzymes; Rolling circle amplification; Structural elements.
© 2025 The Author(s). Angewandte Chemie International Edition published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Colorimetric RT-LAMP for SARS-CoV-2 detection from nasopharyngeal swabs or crude saliva: a multicountry diagnostic accuracy study in Africa.Lancet Glob Health. 2025 Jul;13(7):e1258-e1267. doi: 10.1016/S2214-109X(25)00150-0. Lancet Glob Health. 2025. PMID: 40580991 Free PMC article.
-
Detection of Large Genomic RNA via DNAzyme-Mediated RNA Cleavage and Rolling Circle Amplification: SARS-CoV-2 as a Model.Chemistry. 2023 May 11;29(27):e202300075. doi: 10.1002/chem.202300075. Epub 2023 Mar 27. Chemistry. 2023. PMID: 36790320
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Abdul Wahab M. R., Palaniyandi T., Viswanathan S., Baskar G., Surendran H., Gangadharan S. G. D., Sugumaran A., Sivaji A., Kaliamoorthy S., Kumarasamy S., Clin. Chim. Acta 2024, 555, 117792. - PubMed
-
- Moorthy D. N., Dhinasekaran D., Rebecca P. N. B., Rajendran A. R., J. Biophotonics 2024, 17, e202400243. - PubMed
-
- Sarkar S., Gogoi M., Mahato M., Joshi A. B., Baruah A. J., Kodgire P., Boruah P., Biomed. Microdevices 2022, 24, 32. - PubMed
-
- Senkus E., Kyriakides S., Penault‐Llorca F., Poortmans P. M., Thompson A. J., Zackrisson S., Cardoso F., Ann. Oncol. 2019, 24, 1194–1220. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

