Evolutionary characterization and pathogenicity of a porcine G9P[23] rotavirus with gene segments linked to canine and giant panda strains
- PMID: 40527435
- PMCID: PMC12221699
- DOI: 10.1016/j.virusres.2025.199600
Evolutionary characterization and pathogenicity of a porcine G9P[23] rotavirus with gene segments linked to canine and giant panda strains
Abstract
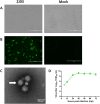
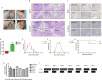
Porcine rotavirus A (RVA) has emerged as an increasingly consequential zoonotic pathogen, causing severe intestinal disorders across diverse mammalian species, including humans. During of an outbreak that struck nursing piglets with diarrhea, a porcine G9P[23] rotavirus, named as RVA/Pig-wt/China/ZJ03/2022/G9P[23] (hereafter referred to as ZJ03), was identified. To further elucidate the evolutionary diversity of ZJ03, a comprehensive analysis of all genome segments was conducted. The genome constellation was identified as G9-P[23]-I5-R1-C1-M1-A8-N1-T1-E1-H1. Nucleotide sequence identity and phylogenetic analyses indicated that the VP3 and NSP1 genes of ZJ03 are most closely related to the corresponding genes of the giant panda strain and the dog strain, respectively, showing the highest homology at 95.73 % identity and 94.64 %. The remaining genes demonstrated the most intimate relationship with porcine strains. Their highest homology levels ranged from 95.98 % to 99.49 % similarity. Therefore, evidence suggests interspecies transmission and genetic reassortment events between porcine, canine, and giant panda rotavirus strains. To evaluate the pathogenicity of ZJ03 strain, we experimentally infected 3-day-old piglets oral inoculation with the PoRV ZJ03 strain at a dose of 2 × 10^5.5 TCID50/ml per piglet. The infection resulted in severe diarrhea in all piglets, which occurred at 48 h post-infection (hpi), accompanied by sustained viral shedding and characteristic small intestinal villous atrophy, indicating significant damage to the intestinal epithelium. In vitro, ZJ03 exhibited efficient replication kinetics in MA104 cells, reaching peak titers of 10^9.25 TCID50/mL at 36 h post-infection. This study reports the first documented case of a novel porcine G9P[23] rotavirus with gene segments linked to canine and giant panda strains in mainland China, characterized by high viral titer and virulence. The findings highlight the emergence of a previously unrecorded RVA strain with significant virological and ecological implications.
Keywords: Genome analysis; Isolation; Pathogenicity; Porcine rotavirus; Whole-genome sequencing.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors affirm that none of the work described in this manuscript could have been influenced by any conflicting financial interests or personal relationships they may have. All authors have approved the manuscript for publication. On behalf of my coauthors, I would like to state that the work described was original research that has never been published before and is not currently being considered for partial or full publication anywhere. The enclosed manuscript has the approval of all listed authors.
Figures



Similar articles
-
Evolutionary characterization and pathogenicity of the highly virulent human-porcine reassortant G9P[23] porcine rotavirus HB05 strain in several Chinese provinces.Front Microbiol. 2025 Mar 14;16:1539905. doi: 10.3389/fmicb.2025.1539905. eCollection 2025. Front Microbiol. 2025. PMID: 40160270 Free PMC article.
-
Isolation and characterization of a G9P[23] porcine rotavirus strain AHFY2022 in China.Microb Pathog. 2024 May;190:106612. doi: 10.1016/j.micpath.2024.106612. Epub 2024 Mar 11. Microb Pathog. 2024. PMID: 38467166
-
Analysis of complete genome sequences of G9P[19] rotavirus strains from human and piglet with diarrhea provides evidence for whole-genome interspecies transmission of nonreassorted porcine rotavirus.Infect Genet Evol. 2017 Jan;47:99-108. doi: 10.1016/j.meegid.2016.11.021. Epub 2016 Nov 26. Infect Genet Evol. 2017. PMID: 27894992
-
Genetic Profile of Rotavirus Type A in Children under 5 Years Old in Africa: A Systematic Review of Prevalence.Viruses. 2024 Feb 3;16(2):243. doi: 10.3390/v16020243. Viruses. 2024. PMID: 38400019 Free PMC article.
-
A systematic review of genetic diversity of human rotavirus circulating in South Korea.Infect Genet Evol. 2014 Dec;28:462-9. doi: 10.1016/j.meegid.2014.08.020. Epub 2014 Sep 12. Infect Genet Evol. 2014. PMID: 25218045
References
-
- Akari Y., Hatazawa R., Kuroki H., Ito H., Negoro M., Tanaka T., Miwa H., Sugiura K., Umemoto M., Tanaka S., Ogawa M., Ito M., Fukuda S., Murata T., Taniguchi K., Suga S., Kamiya H., Nakano T., Taniguchi K., Komoto S. Full genome-based characterization of an Asian G3P[6]human rotavirus strain found in a diarrheic child in Japan: evidence for porcine-to-human zoonotic transmission. Infect. Genet. Evol. 2023;115 - PubMed
-
- Chen D., Zhou L., Tian Y., Wu X., Feng L., Zhang X., Liu Z., Pang S., Kang R., Yu J., Ye Y., Wang H., Yang X. Genetic characterization of a novel G9P[23]rotavirus A strain identified in southwestern China with evidence of a reassortment event between human and porcine strains. Arch. Virol. 2019;164(4):1229–1232. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

