Run-and-tumble dynamics of Escherichia coli is governed by its mechanical properties
- PMID: 40527472
- PMCID: PMC12173490
- DOI: 10.1098/rsif.2025.0035
Run-and-tumble dynamics of Escherichia coli is governed by its mechanical properties
Abstract
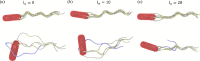
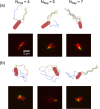
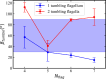
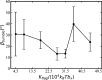
The huge variety of microorganisms motivates fundamental studies of their behaviour with the possibility to construct artificial mimics. A prominent example is the Escherichia coli bacterium, which employs several helical flagella to exhibit a motility pattern that alternates between run (directional swimming) and tumble (change in swimming direction) phases. We establish a detailed E. coli model, coupled to fluid flow described by the dissipative particle dynamics method, and investigate its run-and-tumble behaviour. Different E. coli characteristics, including body geometry, flagella bending rigidity, the number of flagella and their arrangement at the body, are considered. Experiments are also performed to directly compare with the model. Interestingly, in both simulations and experiments, the swimming velocity is nearly independent of the number of flagella. The rigidity of a hook (the short part of a flagellum that connects it directly to the motor), polymorphic transformation (spontaneous change in flagella helicity) of flagella and their arrangement at the body surface strongly influence the run-and-tumble behaviour. Mesoscale hydrodynamics simulations with the developed model help us better understand physical mechanisms that govern E. coli dynamics, yielding the run-and-tumble behaviour that compares well with experimental observations. This model can further be used to explore the behaviour of E. coli and other peritrichous bacteria in more complex realistic environments.
Keywords: bacterium model; hydrodynamic simulation; motility; navigation; propulsion.
Conflict of interest statement
We declare we have no competing interests.
Figures








References
-
- Lauga E, Powers TR. 2009. The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 72, 096601. ( 10.1088/0034-4885/72/9/096601) - DOI
-
- Bechinger C, Di Leonardo R, Löwen H, Reichhardt C, Volpe G, Volpe G. 2016. Active particles in complex and crowded environments. Rev. Mod. Phys. 88, 045006. ( 10.1103/revmodphys.88.045006) - DOI
-
- Gray J, Hancock GJ. 1955. The propulsion of sea-urchin spermatozoa. J. Exp. Biol. 32, 802–814. ( 10.1242/jeb.32.4.802) - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

